
Autophagy is required for PTEN-loss driven prostate cancer
Protein catabolic pathways are of critical importance to cellular physiology. The breakdown of proteins and organelles serves not only as a form of protein and organelle quality control, but also to generate amino acids and free fatty acids for reuse by the cell. Macroautophagy (hereafter referred to as autophagy) is a major pathway of protein degradation in eukaryotic cells that has been preserved through evolution. There has been much interest in the role of autophagy and cancer because mutations in autophagy genes have been observed in human cancers, autophagic activity is altered in cancer cells, and suppression of autophagy could be utilized as a cancer therapy (1,2). A recent report by the DiPaola group (3) demonstrated for the first time that autophagy is essential for the development of prostate cancer driven by PTEN-loss in a mouse model.
Autophagy: a core degradation pathway
Autophagy or “self-eating” is the process, by which long lived cytoplasmic proteins, receptors and organelles are loaded into autophagosomes (double-membrane vesicles), which subsequently fuse with lysosomes for degradation of their cargo by lysosomal enzyme activity. The formation of the autophagosome is an incredibly complex process that has been reviewed in detail by others (4). In brief, formation of the autophagosome occurs in three steps: initiation, nucleation, and expansion/closure (Figure 1). In response to nutrient deprivation, a well-established trigger for autophagy, the Beclin-1 and ULK1 protein kinase complexes are activated and recruited to the endoplasmic reticulum (ER). The Beclin-1 complex is composed of VPS34, a type-III phosphatidylinositol-3 kinase (PI3K), Beclin-1, and ATG14L (4). The ULK complex phosphorylates Beclin-1 to promote translocation of the complex to the ER (5). Once activated, the Beclin-1 complex enriches localized sites on the ER membrane with phosphatidylinositol-3 phosphate (PI3P) via VPS34 activity. These PI3P-enriched domains are critical for nucleation of the isolation membrane or phagophore, a pre-autophagosome structure. The PI3P-enriched phagophore serves as a docking site for WIPI2 (6). The expansion of the autophagosome from the isolation membrane requires the ATG16L1 protein complex by promoting LC3 family protein lipidation and incorporation into the isolation membrane/autophagosome (7). The ATG16L1 complex is recruited to the isolation membrane by interaction with WIPI2, where it promotes LC3 family protein conjugation to ATG3, which promotes phosphatidylethanolamine (PE) lipidation of LC3 (7). The lipidated LC3 proteins are incorporated into the emerging isolation membrane/autophagosome to favor its expansion and closure (8,9). LC3 proteins are also critical for cargo specific loading of autophagosomes via interaction with p62- and NBR1-bound to targets (e.g., mitochondria or protein aggregates) destined for autophagic catabolism (10,11).
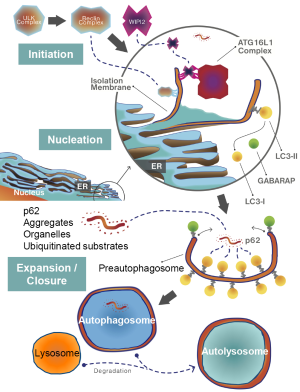
Autophagy and cancer
The relationship between autophagy and cancer is complex. It has been widely observed that deficiencies in core autophagy genes (i.e., genes essential to the autophagy conjugation system) lead to increases in cytoplasmic volume, mitochondria number, and mitochondrial reactive oxygen species (12,13). Subsequently, oxidative DNA damage and DNA double strand breaks are observed in cells lacking core autophagy genes. However, the relationship between autophagy deficiency and cancer is not linear. Where monoallelic loss or down-regulation of core autophagy genes can promote the development of cancer, complete deletion of these same genes can inhibit cancer growth and progression (1,13). It was recently shown that homozygous deletion of ATG7, a regulator of LC3-I conjugation and ATG16L1 complex formation, suppresses melanoma growth by generating high levels of DNA damage and by activating senescence in cancer cells (14). Thus, it could be hypothesized that increased DNA damage as a result of a mild or intermediate defect in autophagy could promote tumorigenesis and drive cancer progression by increasing mutation frequency, whereas complete inhibition of autophagy disrupts organelle and protein quality control mechanisms and generates irreparable DNA damage to trigger senescence and suppress cancer. Direct evidence for the role of monoallelic loss of ATG5 in promoting melanoma progression in human cancer was recently demonstrated (1). This study showed that monoallelic loss of ATG5 in melanoma patient samples was associated with metastatic disease and predicted worsened overall patient survival. The authors also showed that ATG5 haploinsufficiency increased tumor burden and metastasis in a mouse melanoma model driven by PTEN-loss and activated BRAF and promoted resistance to BRAF chemical inhibitors. However, complete ATG5 knockout in this model of melanoma increased sensitivity to BRAF inhibitors and ameliorated tumor burden. Complete loss of ATG7 was also found to delay tumor onset in this same model of melanoma and in a BRAF-driven model of lung cancer (1,14,15). A recent study analyzing single nucleotide polymorphisms (SNP) from 458 patients with localized prostate cancer found that there was an association with biochemical recurrence (BCR) and ATG16L1 SNPs (16). Patients with at least one ATG16L1 rs78835907 A allele showed a 22% reduction in the risk of BCR with an associated increase in ATG16L1 gene expression. Overall, these studies suggest that upregulation of autophagy genes suppresses cancer progression, haploinsufficiency of core autophagy genes promotes cancer progression, and homozygous loss suppresses tumor growth and promotes susceptibility to targeted therapy (Figure 2).
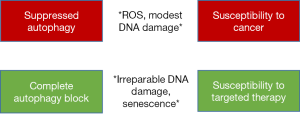
Role of autophagy in PTEN-null prostate cancer
Santanam and colleagues found that loss of ATG7, a core autophagy gene, suppresses prostate tumorigenesis in a PTEN-loss driven mouse prostate cancer model (3). The authors used a novel transgenic mouse with tamoxifen-inducible Cre under control of the Nkx3.1 promoter for inducible expression of Cre in prostate epithelial cells. Prostate specific knockout of PTEN and ATG7 was achieved by administration of tamoxifen to transgenic mice with floxed PTEN and ATG7 alleles. This mouse model of prostate cancer is a significant improvement upon the prior Probasin-Cre mouse because it circumvents the requirement for Cre to be inherited from the male and gives control over PTEN deletion via Cre induction with tamoxifen. As expected, mice with double knockout showed a deficiency in autophagy within their prostate tissue compared to mice harboring PTEN knockout alone as evidenced by increased cytoplasmic volume and accumulation of p62. Concurrent loss of prostatic ATG7 and PTEN resulted in decreased prostate epithelial cell proliferation and apoptotic cell death. Similar to mouse lung and skin cancer models, suppression of autophagy delayed prostate tumor growth and promoted an ER stress response (17). Lastly, the authors castrated control and ATG7 conditional knockout mice and evaluated prostate tumor growth. The ATG7 conditional knockout prostate tumors, also lacking PTEN, showed a greater decrease in prostate volume in response to castration. As the PTEN loss driven prostate cancer model is thought to be castrate-resistant (18,19) it may have been additionally informative to determine whether ATG7 deletion increased the apoptotic response to surgical castration, as these mice showed an increase in ER-stress signaling within their prostate tissue and surgical castration increases apoptosis in prostate cells in this model (18).
Future directions
This study adds to our understanding of the role of autophagy in the PTEN-loss driven mouse prostate cancer model, although several questions still remain. It is unclear whether autophagy is changed in the prostates in mice with prostatic PTEN KO alone relative to benign prostate tissue. PTEN has previously been shown to activate autophagy, so it is possible that loss of PTEN in the prostate may down-regulate autophagy to drive cancer development (20). Further, it is not currently known whether heterozygous loss of ATG7 would promote prostate cancer progression similar to the effect of monoallelic loss of ATG5 in melanoma (1). As the authors suggested, it would be of interest to test whether suppression of autophagy augments response to chemotherapy in this model of prostate cancer. Our own group has found that suppression of autophagy by knockdown of C/EBPβ, a transcriptional regulator of multiple autophagy genes, suppresses the growth of PC-3 xenografts and increased their response to bortezomib (21). ATG7 knockout would be predicted to promote prostate cancer apoptosis in response to docetaxel, as chemical inhibition of autophagy was found to increase docetaxel sensitivity in prostate tumor xenografts (22). Others have shown that blocking autophagy with lysosomotropic agents augments killing of prostate cancer cells by AKT inhibitors (23). Another intriguing possibility is whether the loss of ATG7 increases senescence in PTEN-loss driven prostate cancer as was observed in a mouse model of melanoma (14). Several groups including our own have observed that androgen-deprivation promotes senescence in androgen sensitive prostate cancer cells (24,25). Because senescence occurs in response to loss of PTEN or loss of core autophagy genes, it is possible that ATG7 KO increases the senescence response to PTEN loss and surgical castration as an underlying mechanism for increased sensitivity to androgen deprivation in these mice (26). In conclusion, autophagy plays a critical role in the progression of PTEN-loss driven cancers. Small molecules targeting the autophagy pathway could serve as an adjunct to chemotherapy or androgen deprivation in the treatment of prostate cancer. Future studies aimed at determining the role of autophagy in prostate cancer progression and metastasis are warranted.
Acknowledgments
We would like to thank Michael Barakat for his assistance with preparation of Figure 1.
Funding: This work was supported by the Department of Defense Prostate Cancer Program (W81XWH-14-0209 to DJ Barakat, W81XWH-16-0334 to AD Friedman) by the Patrick C. Walsh Cancer Research Fund (to AD Friedman).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Peng Zhang (Department of Urology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.10.19). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- García-Fernández M, Karras P, Checinska A, et al. Metastatic risk and resistance to BRAF inhibitors in melanoma defined by selective allelic loss of ATG5. Autophagy 2016;12:1776-90. [Crossref] [PubMed]
- Liang XH, Jackson S, Seaman M, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 1999;402:672-6. [Crossref] [PubMed]
- Santanam U, Banach-Petrosky W, Abate-Shen C, et al. Atg7 cooperates with Pten loss to drive prostate cancer tumor growth. Genes Dev 2016;30:399-407. [Crossref] [PubMed]
- Kaur J, Debnath J. Autophagy at the crossroads of catabolism and anabolism. Nat Rev Mol Cell Biol 2015;16:461-72. [Crossref] [PubMed]
- Russell RC, Tian Y, Yuan H, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol 2013;15:741-50. [Crossref] [PubMed]
- Dooley HC, Razi M, Polson HE, et al. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol Cell 2014;55:238-52. [Crossref] [PubMed]
- Fujita N, Itoh T, Omori H, et al. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell 2008;19:2092-100. [Crossref] [PubMed]
- Kabeya Y, Mizushima N, Ueno T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 2000;19:5720-8. [Crossref] [PubMed]
- Weidberg H, Shvets E, Shpilka T, et al. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J 2010;29:1792-802. [Crossref] [PubMed]
- Pankiv S, Clausen TH, Lamark T, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007;282:24131-45. [Crossref] [PubMed]
- Kirkin V, Lamark T, Sou YS, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell 2009;33:505-16. [Crossref] [PubMed]
- Xiong Y, Contento AL, Bassham DC. Disruption of autophagy results in constitutive oxidative stress in Arabidopsis. Autophagy 2007;3:257-8. [Crossref] [PubMed]
- Karantza-Wadsworth V, Patel S, Kravchuk O, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev 2007;21:1621-35. [Crossref] [PubMed]
- Xie X, Koh JY, Price S, et al. Atg7 Overcomes Senescence and Promotes Growth of BrafV600E-Driven Melanoma. Cancer Discov 2015;5:410-23. [Crossref] [PubMed]
- Strohecker AM, Guo JY, Karsli-Uzunbas G, et al. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer Discov 2013;3:1272-85. [Crossref] [PubMed]
- Huang CY, Huang SP, Lin VC, et al. Genetic variants of the autophagy pathway as prognostic indicators for prostate cancer. Sci Rep 2015;5:14045. [Crossref] [PubMed]
- Ma XH, Piao SF, Dey S, et al. Targeting ER stress-induced autophagy overcomes BRAF inhibitor resistance in melanoma. J Clin Invest 2014;124:1406-17. [Crossref] [PubMed]
- Wang S, Gao J, Lei Q, et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell 2003;4:209-21. [Crossref] [PubMed]
- Mulholland DJ, Tran LM, Li Y, et al. Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell 2011;19:792-804. [Crossref] [PubMed]
- Ueno T, Sato W, Horie Y, et al. Loss of Pten, a tumor suppressor, causes the strong inhibition of autophagy without affecting LC3 lipidation. Autophagy 2008;4:692-700. [Crossref] [PubMed]
- Barakat DJ, Mendonca J, Barberi T, et al. C/EBPβ regulates sensitivity to bortezomib in prostate cancer cells by inducing REDD1 and autophagosome-lysosome fusion. Cancer Lett 2016;375:152-61. [Crossref] [PubMed]
- Tan Q, Joshua AM, Saggar JK, et al. Effect of pantoprazole to enhance activity of docetaxel against human tumour xenografts by inhibiting autophagy. Br J Cancer 2015;112:832-40. [Crossref] [PubMed]
- Lamoureux F, Thomas C, Crafter C, et al. Blocked autophagy using lysosomotropic agents sensitizes resistant prostate tumor cells to the novel Akt inhibitor AZD5363. Clin Cancer Res 2013;19:833-44. [Crossref] [PubMed]
- Barakat DJ, Zhang J, Barberi T, et al. CCAAT/Enhancer binding protein β controls androgen-deprivation-induced senescence in prostate cancer cells. Oncogene 2015;34:5912-22. [Crossref] [PubMed]
- Ewald JA, Desotelle JA, Church DR, et al. Androgen deprivation induces senescence characteristics in prostate cancer cells in vitro and in vivo. Prostate 2013;73:337-45. [Crossref] [PubMed]
- Chen Z, Trotman LC, Shaffer D, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 2005;436:725-30. [Crossref] [PubMed]


