
Challenges and opportunities of CYP3A5 as novel drug target in pancreatic cancer subtypes
We thank both Andrew J. Scott and John C. Wilkinson for their excellent commentary about our paper and their stimulating remarks. We would like to take this opportunity to comment on some of these and further discuss some aspects of our data in greater depth than was possible in the original manuscript (1). The marker classification as described by Scott and Wilkinson is somewhat simplified compared to the one we used (1). We only used the PDAssigner described by Collisson et al. (2) for our PACO cell lines and respective xenografts as for these we had transcriptional profiles available. Immunohistochemical samples of the PDAC patient cohort were classified with the surrogate markers HNF1A and KRT81, and thus the samples were designated accordingly. While there is significant overlap between the stratification of patients using our markers and the PDAssigner, the match is not perfect (Noll et al., supplementary image 1F) (1). One reason could be the fact that some genes of the PDAssigner are also expressed by normal pancreas (2). This suggests a potential contamination of the laser micro-dissected epithelial tumor cells used to derive the PDAssigner with normal tissue. Even though one should not exclude the possibility that tumors express genes present in the normal epithelium, refinement of the PDAssigner might further improve stratification of PDAC subtypes based on transcriptional profiling.
Moffitt et al. described an alternative method for PDAC classification using a more stringent filtering approach removing genes highly expressed in normal tissues resulting in two distinct PDAC subtypes (3). Bailey et al. used transcriptional profiling on a large cohort of PDAC samples, identifying four subtypes that were shown to correspond in part to the subtypes described by Collisson et al. (2,4). One of the most consistent signatures across all studies is the one for the quasimesenchymal (QM-PDA) subtype, also termed basal [Bailey et al. (4)] or squamous [Moffitt et al. (3)], respectively. The integration of these recently described classification algorithms with the PDAssigner signature may significantly advance PDAC subtype stratification of independent gene expression datasets. Moreover, putative associations of our marker-defined PDAC subtypes with the subtypes defined by Moffitt et al. and Bailey et al. should be evaluated to further advance routine PDAC subtype stratification at RNA and protein level.
Of note, we identified some HNF1A/KRT81 double positive cases in our tissue microarray analysis (1), suggesting a significant degree of tumor heterogeneity and plasticity between the subtypes with potential implications for PDAC classification and response to therapy (1). Thus, our staining protocol and evaluation algorithm for PDAC stratification according to the expression of KRT81 and HNF1A must be further independently validated using additional patient cohorts and in prospective studies before it should enter into routine diagnostics.
With regard to Table 1 of the editorial, we would like to point out that the stated mean survival reflects the survival of the 10–20% of PDAC patients with resectable tumors and thus localized disease and inherently better prognosis. The survival of the majority of PDAC patients is significantly lower (5). Furthermore, while most HNF1A+ cases were indeed CYP3A5+, this is not the case for all HNF1A+ specimens (Table 1). We also identified CYP3A5 positive double negative (DN) (classical) and KRT81+ PDAC cases. Since our data mechanistically link CYP3A5 expression to drug resistance, its expression should be taken into consideration independently of subtype stratification when interpreting clinical studies using novel PDAC drug treatments.
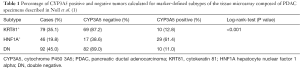
Full table
Scott and Wilkinson mention that systemic CYP inhibition in combination with chemotherapy could be problematic in a therapeutic setting due to the expected toxicity. However, while this may be true for pan-CYP inhibitors, specific inhibition of CYP3A5 may offer significant therapeutic potential. Carriers of the CYP3A5*3, CYP3A5*5 and CYP3A5*6 polymorphisms in the human population show low or absent expression of CYP3A5 without any apparent phenotype, suggesting that CYP3A5 itself is dispensable for normal physiology (6). However, it may be challenging to design such a specific inhibitor owing to structural similarities to the other CYP3A family members (6). Importantly, a putative therapeutic window could be established using CYP3A family inhibitors in combination with small molecule PDAC drugs to boost the intracellular concentration of those within the PDAC cells while side effects remain tolerable.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Xiaotian Sun (Department of Internal Medicine Clinic of August First Film Studio, Beijing, China).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.10.59). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Noll EM, Eisen C, Stenzinger A, et al. CYP3A5 mediates basal and acquired therapy resistance in different subtypes of pancreatic ductal adenocarcinoma. Nat Med 2016;22:278-87. [Crossref] [PubMed]
- Collisson EA, Sadanandam A, Olson P, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med 2011;17:500-3. [Crossref] [PubMed]
- Moffitt RA, Marayati R, Flate EL, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet 2015;47:1168-78. [Crossref] [PubMed]
- Bailey P, Chang DK, Nones K, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531:47-52. [Crossref] [PubMed]
- Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med 2014;371:1039-49. [Crossref] [PubMed]
- Westlind-Johnsson A, Malmebo S, Johansson A, et al. Comparative analysis of CYP3A expression in human liver suggests only a minor role for CYP3A5 in drug metabolism. Drug Metab Dispos 2003;31:755-61. [Crossref] [PubMed]


