
Genetic and epigenetic alterations in urothelial carcinoma
Urothelial carcinoma of the bladder (UCB) is the second most common malignancy of the genitourinary tract and one of the major cancers worldwide (1). In clinical practice, UCB can be roughly classified into non-muscle-invasive bladder cancer (NMIBC) (i.e., Ta, Tis, and T1) and muscle-invasive bladder cancer (MIBC) (i.e., ≥ T2 tumors). At initial presentation, approximately 75% of the UCB tumors are diagnosed with NMIBC; however, 50–70% of them will recur and progress to MIBC (2). Thus, exploring the molecular mechanisms underpinning the development and progression of UCB are important for both accurate diagnosis and precise treatment.
Genomic and epigenomic studies clearly uncovered that significant variations are noted among NMIBC or MIBC tumors, leading to aggressive investigation in the field of molecular sub-grouping in UBC (3), mainly focusing on MIBC (4). In this regard, two main molecular subtypes are found: luminal- and basal-type tumors. Of these, papillary tumors have been reported to correlate with luminal type; on the other hand, non-papillary tumors [including carcinoma in situ (CIS)] are associated with basal type (3). Remarkably, these molecular subtypes have been reported to correlate with several clinical endpoints, such as intrinsic chemosensitivity and patient outcomes. However, till now, investigation for molecularly subtyping NMIBC is limited.
Recently, by using next-generation sequencing (NGS) platform to conduct total RNA sequencing in 460 European NMIBC patients, Hedegaard et al. successfully sub-grouped NMIBC into three molecular classes according to luminal- and basal-like features (5). Varied biological processes, mutation signatures, and clinical outcomes are identified among the three molecular classes, implicating different treatment recommendation.
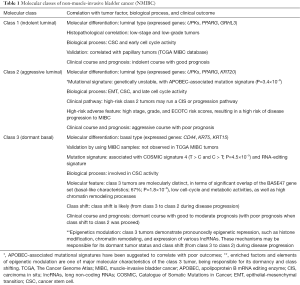
First, in the discovery cohort, Hedegaard et al. used unsupervised consensus clustering (6) to classify 8,074 genes for identifying class 1, 2, and 3 tumors. The three molecular classes are not only differed in clinical-histopathological features, but also showed diversely clinical outcomes (Table 1). For example, when compared with class 1 tumors, tumors of class 2 and class 3 demonstrated aggressive profiles of higher cancer stage, histopathological grade, and risk of progression to MIBC.
Full table
In molecular profiles, class 1 tumors present luminal-like features with a high expression of early cell-cycle genes, demonstrating an indolent disease course and a good prognosis. In contrast, class 2 tumors show a different luminal-like molecular pattern with high expression of late cell-cycle genes, association with cancer aggressiveness and disease progression, behaving poor clinical outcomes.
Interestingly, by using total RNA sequencing to include survey of non-coding RNAs, the authors firstly identified class 3 tumors in early-stage NMIBC bladder cancer. Note that this investigating strategy is different from the The Cancer Genome Atlas (TCGA) and several microarray dataset (e.g., the Lund set); both of them allocate protein-coding genes only. Remarkably, the class 3 tumors demonstrate several unique roles in both biological and clinical aspects (Table 1). For example, by using previously published molecular subtyping panels for validation, the authors observed that the basal-like BASE47 MIBC signature (4) is significantly overlapped with class 3 tumors (67%; chi-square test, P=1.8×10−9). This finding is also observed firstly in NMIBC, gaining an interest for further investigation.
For the newly identified class 3 tumors, it is noteworthy to further point out that genes involved in epigenetic modifications play an important role. For example, repressed-gene-enrich profile and high induced histone/chromatin modifications are observed over targets of several transcription factors. More notably, these class 3 basal-like tumors also demonstrate pronounced expression of long non-coding RNAs (lncRNAs). Epigenetic modifications including DNA methylation, histone modifications and expression of non-coding RNAs are important mechanisms in regulating gene expression. Although this study provides a comprehensive expression profile of histone/chromatin modifiers and lncRNAs, an integrated transcriptomic and epigenomic profile in bladder cancer is still lacking. Several studies including ours have investigated the genome-wide DNA methylation profile in bladder cancer either by microarray or NGS platform (7-10). Of noted, one study used NGS platform to investigate the integrative DNA methylation and the expression of both mRNA and miRNAs in nine primary bladder cancer tissue samples, confirming that pathways related to neurogenesis and cell differentiation are enriched in those patient samples (8). Recently, by using methylation microarray, we also found that methylation of ZNF671 is associated with recurrent bladder cancer patients and poor locoregional disease-free survival (7). Integrative information on transcriptomic, genomic and epigenomic profile will not only provide information regarding the carcinogenesis of bladder cancer but also be able to identify biomarkers or signatures for accurate disease diagnosis, outcome prediction, and treatment selection.
Finally, Hedegaard et al. identified six distinct trinucleotide mutational signatures. Interestingly, they found that one of the signatures (signature 3) closely resembled the apolipoprotein B mRNA editing enzyme (APOBEC)-associated mutational signature. Importantly, aggressive class 2 tumor is predominantly associated with this signature probably due to overexpression of APOBEC3A and APOBEC3B in this tumor subtype. APOBEC is a family of evolutionally conserved proteins involving in mRNA editing processes. The APOBEC protein family contains the catalytic cytidine deaminase domain for C-to-U mRNA editing processes. Similarly, the Catalogue of Somatic Mutations in Cancer (COSMIC) database, which is the most comprehensive online resource for somatic mutations of human cancers, has so far reported a total of 30 mutation signatures (11). Of these, two signatures (signature 2 and 13) were related to APOBEC. Recently studies also found that, APOBEC-associated signature is correlated with poor prognosis in multiple myeloma (7) and urothelial carcinoma (8).
Clinically, though several limitations do exist, these results, together with data from a prior study (12), make liquid biopsy (either blood or urine) closer for clinical application in patients with urothelial carcinoma. Biologically, the author declared two interesting pathways that may be useful to demarcate the processes of urothelial carcinogenesis: the Ta (class 1 and 3) and the CIS pathway (class 2). In this regard, class 1 and 2 tumors are able to progress to MIBC by different pathways. On the other hand, class 3 tumors that initially conduct the Ta pathway (or under a dormant status) are able to execute class shift to class 2 and then progress to MIBC.
In summary, three take-home messages are noted. First, after validation in independent cohorts, the authors successfully validated a concise 117-gene panel that is efficient to conduct molecular subtyping, increasing the possibility of clinical application. Second, the three molecular classes well correlate with progression-free survival: class 1 (the best prognosis) > class 3 > class 2 tumors (the poorest outcomes). Remarkably, third, class 3 tumors characterized pronouncedly epigenetically modified disease profiles, implicated a role of epigenetic intervention. Further multi-center prospective studies are of large interest and warranted.
Acknowledgments
Funding: The present work was supported by Buddhist Dalin Tzu Chi Hospital and Ministry of Science and Technology, Taiwan (MOST 104-2314-B-194-001-MY3).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Peng Zhang (Department of Urology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.11.47). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7-30. [Crossref] [PubMed]
- Halperin EC, Brady LW, Wazer DE, et al. editors. Perez and Brady's principles and practice of radiation oncology. 6th edition. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins, 2013.
- Czerniak B, Dinney C, McConkey D. Origins of Bladder Cancer. Annu Rev Pathol 2016;11:149-74. [Crossref] [PubMed]
- Damrauer JS, Hoadley KA, Chism DD, et al. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proc Natl Acad Sci U S A 2014;111:3110-5. [Crossref] [PubMed]
- Hedegaard J, Lamy P, Nordentoft I, et al. Comprehensive Transcriptional Analysis of Early-Stage Urothelial Carcinoma. Cancer Cell 2016;30:27-42. [Crossref] [PubMed]
- Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 2010;26:1572-3. [Crossref] [PubMed]
- Yeh CM, Chen PC, Hsieh HY, et al. Methylomics analysis identifies ZNF671 as an epigenetically repressed novel tumor suppressor and a potential non-invasive biomarker for the detection of urothelial carcinoma. Oncotarget 2015;6:29555-72. [PubMed]
- Zhu J, Jiang Z, Gao F, et al. A systematic analysis on DNA methylation and the expression of both mRNA and microRNA in bladder cancer. PLoS One 2011;6:e28223 [Crossref] [PubMed]
- Kandimalla R, van Tilborg AA, Kompier LC, et al. Genome-wide analysis of CpG island methylation in bladder cancer identified TBX2, TBX3, GATA2, and ZIC4 as pTa-specific prognostic markers. Eur Urol 2012;61:1245-56. [Crossref] [PubMed]
- Ibragimova I, Dulaimi E, Slifker MJ, et al. A global profile of gene promoter methylation in treatment-naïve urothelial cancer. Epigenetics 2014;9:760-73. [Crossref] [PubMed]
- COSMIC, Catalogue of Somatic Mutations in Cancer. Signatures of Mutational Processes in Human Cancer. Available online: http://cancer.sanger.ac.uk/cosmic/signatures
- Birkenkamp-Demtröder K, Nordentoft I, Christensen E, et al. Genomic Alterations in Liquid Biopsies from Patients with Bladder Cancer. Eur Urol 2016;70:75-82. [Crossref] [PubMed]


