
In vivo cancer metabolism is defined by the nutrient microenvironment
Cancer cells show sustained cellular proliferation, which is necessary for tumor formation and cancer progression. Cellular metabolism is one of the biological processes necessary to enable uncontrolled proliferation of cancer cells. Metabolism converts nutrients provided by the microenvironment through a series of biochemical reactions into metabolic products such as energy, biomass precursors, and antioxidants, which are in turn needed for cellular proliferation. Yet, metabolism can be fueled by different nutrients, and these nutrients are used to produce the needed metabolic products through multiple parallel or converging biochemical reactions. Thus, almost a century ago the idea emerged that cancer cells have a different metabolism compared to non-transformed cells (1) and that these differences can be targeted to inhibit cancer cell proliferation (2).
After the challenging discovery that there is no specific “cancer metabolism” that unifies all transformed cells and separates them from all non-transformed cells (3), researchers undertook the endeavor to mechanistically understand how metabolic alterations are linked to an oncogenic transformation. A milestone in this endeavor was the discovery that oncogenes and tumor suppressors regulate the expression and/or activity of metabolic enzymes (3,4). Consequently, it has been concluded that cancer metabolism is defined by the genetic landscape of the transformed cell. Yet, introducing one and the same oncogenic driver into different non-transformed cells often did not result in a consistent change in metabolism (5-7). This finding was explained by the frequently underappreciated influence of the cell origin (3), i.e., non-transformed cells of different origin have per se a different metabolism, which is defined by their natural function within an organ or the whole body. Thus, the metabolic changes stimulated by the introduction of an oncogenic driver in cells of different origin (and therefore different baseline metabolism), result in a metabolism that is defined by both, the oncogenic drivers and the cell origin (3). Yet, with these findings the question rises whether cancer metabolism is only defined by cell intrinsic factors such as the genetic landscape and cell origin. Multiple lines of evidence, such as metabolic regulation and the importance of extracellular nutrient concentrations on the activity of signaling pathways (8), suggest otherwise. However, the impact of these cell extrinsic factors has hardly been studied in vivo. Consequently, the relevance and interplay between cell intrinsic and extrinsic factors in shaping in vivo cancer metabolism is poorly defined.
Recently, several laboratories addressed the question whether cell extrinsic factors such as alterations in the microenvironment, play a role in defining cancer metabolism. Hensley and colleagues (9) studied the interplay between metabolism and the microenvironment by combining in vivo 13C tracer analysis with dynamic contrast-enhanced MRI (DCE-MRI) data in non-small cell lung cancer patients with differential status of the oncogenes KRAS and EGFR. Using DCE-MRI they pre-operatively defined higher and lower perfused areas within the cancer mass and used this information to identify cancer cells exposed to different microenvironments, although harboring the same oncogenic drivers and residing in the same organ. The consequent intraoperative 13C tracer analysis was used to determine differential nutrient usage and metabolic pathway activities (10) of the cancer cells located in different microenvironments (defined by the perfusion magnitude) within the cancer mass. Based on this data they came to three conclusions (Figure 1): first, regardless of the perfusion state all cancer cells showed sustained or even enhanced tricarboxylic acid (TCA) cycle metabolism [defined by pyruvate carboxylase (PC)-dependent anaplerosis and pyruvate dehydrogenase (PDH) activity] despite the fact that they also showed increased glucose uptake (measured by FDG-PET) compared to adjacent lung tissue. This finding is of profound importance, because a long favored model of cancer metabolism postulates a switch from oxidative to glycolytic metabolism. However, the data of Hensley and colleagues (9) support the idea that enhanced oxidative TCA cycle metabolism and increased glycolysis are not opposing metabolic states, but can both be the metabolic basis of an oncogenic transformation; second, cancer cells in lower perfused areas within the cancer mass relied predominantly on glucose to fuel TCA cycle metabolism, while cancer cells in areas of higher perfusion used to a large extent alternative carbon sources; third, neither the KRAS nor the EGFR status changed the influence of perfusion on the nutrient usage in the TCA cycle.
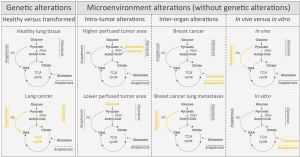
These unexpected results were further confirmed and extended by a study of Davidson and colleagues (6) showing that cellular proliferation of KRAS driven non-small cell lung cancers in mice depends in vivo on PC-dependent anaplerosis and PDH activity. Moreover, they discovered that KRAS-driven lung cancer cells used in vivo glucose as major carbon substrate for TCA cycle fueling, while the same cells cultured in vitro switched to glutamine anaplerosis (Figure 1).
Beyond these works on primary lung cancer, Christen and colleagues (11) investigated the role of the microenvironment in shaping cancer metabolism during breast cancer metastasis to the lungs. They discovered that breast cancer-derived lung metastases activated PC-dependent anaplerosis as a function of the nutrient availability within the lung microenvironment (Figure 1). While primary breast cancers often rely on glutamine anaplerosis and are susceptible to glutaminase inhibitors (3), these data suggest that the resulting and genetically similar lung metastases loose this drug susceptibility, because they activate PC-dependent anaplerosis in response to the lung microenvironment (11).
Considering the data provided by these studies three major milestone conclusions emerge: first, cancer metabolism is a function of the in vivo microenvironment resulting in intra-tumor and inter-organ heterogeneity; second, the nutrient microenvironment can overrule metabolic constrains imposed by the genetic landscape; third, a true switch from oxidative to glycolytic metabolism is not a requirement for uncontrolled proliferation of cancer cells.
At first sight the discovered metabolic heterogeneity and flexibility of cancer cells resulting from the microenvironment seems challenging for the overarching goal to translate cancer metabolism research into innovative and effective drugs against cancer. However, a closer look at these results reveals that although cancers within the lung show the flexibility to exchange glucose with alternative carbon sources, the metabolic processing of all these nutrients requires PC and PDH activity (9). Consequently, targeting PC (or PDH) activity has the potential to inhibit the in vivo proliferation of cancer cells in the lung microenvironment regardless of whether they are located in higher or lesser perfused areas (9), or originate from primary breast cancers (11). However, this in vivo vulnerability of cancer cells within the lung microenvironment is not predictable from in vitro studies, because the in vitro provided environment results in a glutamine dependency of lung cancer cells (6). Thus, these findings provide the basis for a paradigm shift, which builds on a microenvironment defined cancer metabolism. Consequently, not only the genetic landscape and the cell origin of cancer cells needs to be considered when developing anti-cancer drugs and defining their efficacy spectrum, but also the microenvironment as defined by the perfusion state of the tumor, the organ in which the cancer resides, and the whole body physiology of the patient.
Acknowledgments
We thank Dr. Stefan Christen (VIB Leuven) for feedback of the perspective.
Funding: SMF acknowledges funding support from Marie Curie–CIG, FWO–Odysseus II, Concern Foundation, FWO–Research Grants, Eugène Yourassowsky Schenking, KU Leuven–Methusalem Co-Funding, and Bayer Health Care.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Shaohua Cui (Department of Pulmonary Medicine, Shanghai Chest Hospital, Shanghai Jiao Tong University, Shanghai, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.11.53). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Warburg O. The Metabolism of Carcinoma Cells. The Journal of Cancer Research 1925;9:148-63. [Crossref]
- Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol 1927;8:519-30. [Crossref] [PubMed]
- Elia I, Schmieder R, Christen S, et al. Organ-Specific Cancer Metabolism and Its Potential for Therapy. Handb Exp Pharmacol 2016;233:321-53. [Crossref] [PubMed]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 2009;324:1029-33. [Crossref] [PubMed]
- Yuneva MO, Fan TW, Allen TD, et al. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab 2012;15:157-70. [Crossref] [PubMed]
- Davidson SM, Papagiannakopoulos T, Olenchock BA, et al. Environment Impacts the Metabolic Dependencies of Ras-Driven Non-Small Cell Lung Cancer. Cell Metab 2016;23:517-28. [Crossref] [PubMed]
- Mayers JR, Torrence ME, Danai LV, et al. Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science 2016;353:1161-5. [Crossref] [PubMed]
- Lorendeau D, Christen S, Rinaldi G, et al. Metabolic control of signalling pathways and metabolic auto-regulation. Biol Cell 2015;107:251-72. [Crossref] [PubMed]
- Hensley CT, Faubert B, Yuan Q, et al. Metabolic Heterogeneity in Human Lung Tumors. Cell 2016;164:681-94. [Crossref] [PubMed]
- Buescher JM, Antoniewicz MR, Boros LG, et al. A roadmap for interpreting (13)C metabolite labeling patterns from cells. Curr Opin Biotechnol 2015;34:189-201. [Crossref] [PubMed]
- Christen S, Lorendeau D, Schmieder R, et al. Breast Cancer-Derived Lung Metastases Show Increased Pyruvate Carboxylase-Dependent Anaplerosis. Cell Rep 2016;17:837-48. [Crossref] [PubMed]


