
Progress in studies on autoantibodies against tumor-associated antigens in hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common malignant tumor with the third highest mortality in the world. Most HCC patients died within one year after the definitive diagnosis, and the part of the reason is due to lack of effective early diagnosis technology. China has a high incidence rate of HCC and about 50.5% new patients and 51.4% cancer related deaths globally occurred in China each year (1,2). The mechanisms underlying HCC occurrence and development have not been well clarified. Alpha-fetoprotein (AFP), which has been widely used as a diagnostic marker of HCC, has a low sensitivity in some patients, especially for patients with small size, early stage and well differentiated, which cannot meet the clinical requirement for early HCC diagnosis (3,4). Therefore, it is urgent to find reliable HCC markers with high sensitivity and specificity.
As early as 1960, W Baldwin Robert has demonstrated that the immune system could react with tumor cells during their development (5). When normal cells transform into tumor cells, body’s immune surveillance function can detect the abnormal substances in cancer cells, or tumor associated antigens (TAAs). TAAs are proteins, nucleic acids, carbohydrates or immune molecule complexes with antigenic properties abnormally expressed in tumor cells (6). Detection of some tumors can be achieved by identification of their specific TAAs and homologous autoantibodies. The mechanisms for production of anti-TAAs autoantibodies are not clear. Some scholars believed that over-expression, degradation, dislocation and folding of TAAs can induce immune system to produce immune responses, especially humoral immunity. Therefore, their corresponding autoantibodies can be tested in tumor tissues and serum (7). Many studies have reported that the immune responses were mediated by autoantibodies in the development of HCC. Belousov et al. summarized the studies about antigens and autoantibodies associated with prostate cancer, lung cancer, breast cancer, nerve glioma, liver cancer using different detection methods (6) and showed that anti-TAAs autoantibodies have been widely used in the diagnosis and detection of many cancers. Dai et al. have introduced the studies of anti-TAAs autoantibodies and TAAs in HCC since 1993 (8). Some studies have shown that anti-TAAs autoantibodies are more stable than TAAs and not easy to be hydrolyzed (2) and can be used as the “reporter” of the immune system to identify the changes of body’s antigen substances (8). Zhang and Tan proposed that anti-TAAs autoantibodies should be divided into two types, one is the autoantibodies compounded after appearance of tumor cells; the other is continuous presence of autoantibodies before or after tumor appearance (9). Some of anti-TAAs autoantibodies can be used as potential markers or targets for immunotherapy (10,11). However, the role of anti-TAAs autoantibodies of HCC is still poorly understood. This review focuses on the four main HCC screening techniques and the recent progress about anti-TAAs autoantibodies in HCC detection.
Screening techniques of autoantibodies in the diagnosis of HCC
The screen techniques of autoantibodies or TAAs in tissues or serum of HCC patients can be mainly classified as serum proteomics analyses (SERPA), serological analysis of recombinant expressed cDNA clone (SEREX), phage display technology and high throughput protein microarray. These techniques have been proven to be effective. Table 1 summarized the methodologies in the screening of autoantibodies for the diagnosis of HCC. Lots of candidate antigens or autoantibodies have been identified using these techniques and gradually applied in practice.
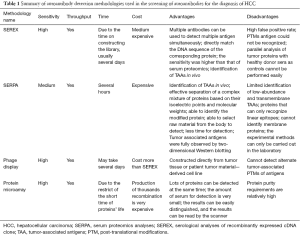
Full table
SERPA
Desmetz et al. has mentioned that “the SERPA technique, also known as PROTEOMEX, has, for several decades, remained one of the best tools for identifying TAAs” (5). SERPA is a technology combination of two-dimensional protein electrophoresis (2-DE), western blot and mass spectrometry analysis. SERPA can detect slight differences in expression of proteins in different cells. The process of SERPA includes several steps (12). Firstly, dissolve cancer and normal cells (tissues), extract total proteins, conduct two-dimensional gel electrophoresis and identify proteins with differential expression in cancer and normal cells (tissues). Secondly, screen different protein loci and analyze the candidate proteins by mass spectrometry. Thirdly, verify their expression in serum using western blot or enzyme linked immunosorbent assay (ELISA). Takashima et al. identified antigens HSP70, GAPDH, PRX and Mn-SOD using SERPA in 2006 and used these antigens to detect the corresponding autoantibodies in serum. They demonstrated that the sensitivity of TAAs is highly correlated with the autoantibodies (13). Looi et al. performed similar experiments (SERPA) in 20 HCC patients and 50 patients with chronic hepatitis (CH) and liver cirrhosis (LC), identified HSP60 and HSP70 as ATTs and verified their results using western blot (14,15). Li et al. discovered 13 different loci and verified 8 of them (Keratin 8, lamin A/C, DEAD, eEF2, hnRNP A2/B1, prostate binding protein and TIM) using western blot. They also found that the sensitivity of serum autoantibodies combined with AFP was 69.8–82.6% for HCC patients (16). Hong et al. performed SERPA of serum samples and identified 20 TAAs. Among these TAAs, autoantibodies of CENPF, HSP60 and IMP-2 have high sensitivity for early HCC patients, reaching 79.3% when combined with AFP (1). Although SERPA technology is very reliable, it has many shortcomings and requires other techniques in screening application. For example, SERPA cannot detect low-abundance, transmembrane proteins and space epitopes of antigen-antibody reaction (17).
Serological analyses of recombinant cDNA expression libraries (SEREX)
SEREX is a combination of serum analysis and molecular genetic recombination technology. It uses recombinant cDNA expression libraries to express recombinant proteins that can react with the serum of cancer patients. The technology was first reported by Undschuh et al. in 1995. Its key step is to establish an effective and comprehensive cDNA expression library (18). The first step to explore the differential autoantibodies is synthesis of protein (TAAs) and the second step is making TAAs react with autoantibodies in the serum samples. By using SEREX technology, high expression of anti-TAAs autoantibodies would be identified according to the reaction with the synthesized TAAs (19). Song et al. successfully reproduced the CT antigens (AKAP3, CTp11) in HCC patients and verified their expression in multiple cancer cells (20). Chen et al. identified Sui1 and RalA antigens from HCC patients using SEREX, and detected their autoantibodies in patients’ sera and showed that the sensitivity of single Sui1 and RalA autoantibodies was 11.7% (9/77) and 19.5% (15/77), respectively, which increased to 66.2% (51/77) when combined with eight known autoantibodies and can even reach 88.7% when combined with AFP in HCC patients (21). Wang et al. separated 81 kinds of antigens from the HepG2 cell line and identified five differential autoantibodies in HCC patients. The individual sensitivity of these autoantibodies was 76.4–91.0% and the sensitivity and specificity when combined with KRT23, AHSG and FTL could reach 98.2%, 90%, respectively (18). SEREX is a high throughput screening technology, but it could not mark off the modified proteins such as glycerogelatin antigen. In addition, difficulties in selecting differentially expressed proteins and long period for preparing expression plasmids, which is not so accurate, have restricted the development of SEREX (5,17).
Phage display technologies
Phage display technology is a technology that uses molecular biology methods to introduce genes which produce exogenous polypeptides or proteins into the genome of phage capsid proteins to express fusion proteins on the surface of phage (22). This method was first reported by Smith et al. in 1985 for displaying polypeptide protein on the surface of filamentous phage (23). Phage display technology is a third generation technology to identify autoantibodies. Depending on the host cells and the expression vector, it can be divided into three types: the filamentous phage display system, T7 or T4 phage display system and λ phage display system. These three types of phage display systems can carry different amounts of proteins, thus they are more advantageous than SEREX technology (24). Talwar et al. used T7 phage display library to screen tuberculosis and sarcoidosis autoantibodies (25). Zhang found that anti-LMP1 autoantibodies were associated with EB virus (26). Zhang utilized filamentous M13 phage display system and combined the screening result with AFP to diagnose patients with early HCC (22). Liu reverse transcribed all mRNAs of HCC cells into cDNA and constructed a cDNA expression library using T7 phage display technology. Liu identified DDX3, CENPF and other tumor-associated antigens (TAAs) after five rounds of serum screening of ten cases of liver cancer and ten cases of healthy and identified DDX3, HSPA4, CENPF, HSPA5, VIM, LMNB1 and TP53 as markers for early diagnosis of HCC by performing ELISA of 70 HCC patients, 50 CH patients and 70 Normal Healthy Serum (NHS) (27). This technique is similar to SEREX, which could express modified proteins and the proteins cannot be expressed as phage coat proteins. Besides, after many rounds of screening, autoantibodies with lower affinity are easy to be lost (2,5,9,17).
Protein microarray and high throughput protein microarray
The protein microarray is based on high-throughput protein chips, and there are thousands of known proteins fixed on it. With the development of molecular genomics, some known protein encoding genes can be expressed in vitro (28). These known recombinant proteins are labeled with special tags and immobilized on the same two chips. The results of protein microarray indicated different expression of autoantibodies which response to assigned protein antigens in the sera samples of patients and healthy control. High throughput protein chips provide a new way for the search of TAAs or autoantibodies. The technology has advantages of high throughput, high automation degree and fast speed. The second generation chip contains nearly 20,000 recombinant proteins (29-32). Although the second generation high-throughput protein chip has been applied for disease diagnosis, it has not been reported in HCC. Hu identified autoantibodies of KLHL12 and ZBTB2 using high throughput protein chip technology and used them as primary markers for diagnosis of biliary cirrhosis (31). Jeong et al. used this chip to rapidly identify specific autoantibodies or antigens (30). All these reports suggest that high-throughput protein chips can be used to screen HCC autoantibodies.
Current researches on the diagnostic autoantibodies in HCC
Although autoantibodies have been identified for long time, they have not been incorporated into diagnostic criteria of cancers. For HCC, single autoantibody has high specificity and low sensitivity, which cannot meet the diagnostic requirements. However, combined autoantibodies are considered could improve these autoantibodies’ diagnosis value of HCC. And many researchers have tested the combined autoantibodies in large samples.
Table 2 lists the latest research advances on single-autoantibodies and the panel of autoantibodies.
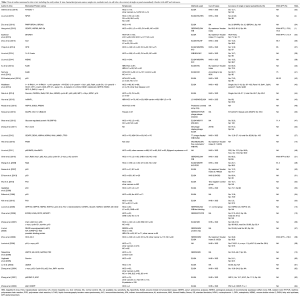
Full table
Autoantibodies for HCC detection
p53, p16, p62, p90
The P53 gene, mapped to chromosome 17p13, encodes a 53-kDa nuclear phosphoprotein (p53). It is involved in the regulation of cell growth, DNA repair, and apoptosis function (50-52). Inactivation of the wild-type p53 is considered to play a key role in the carcinogenesis of many malignancies. . Studies have shown that p53 protein is expressed in many cancers, including lung cancer, esophageal cancer, oral cancer, colon cancer and gastric cancer (8). At present, most researches on autoantibodies are focused on the anti-p53 autoantibodies, a tumor suppressor protein. The sensitivity and specificity of anti-p53 autoantibodies were reported to be 5.3–73.3% and >95% for HCC (21,27,42-44,50-54,65-69). Many studies have shown that the sensitivity of anti-p53 autoantibodies for HCC varied greatly possibly due to the heterogeneity of HCC. For example, EI-Emshaty et al. detected anti-p53 autoantibodies in HCV-HCC in 2014 and showed that its ELISA sensitivity was 73.3% (42). Gadelhak et al. detected anti-p53 autoantibodies and showed its ELISA sensitivity was 73.07% (53). However, Edis et al. and Raedle et al. showed the sensitivity of anti-p53 autoantibodies were only 12.8% and 22.7% (54,70). The sensitivities of anti-p53 autoantibodies varied greatly for HCV-HCC and hepatitis B virus (HBV)-HCC. The former has high sensitivity (42) while the latter has low sensitivity (27,44,52), which suggests that anti-p53 autoantibodies may become a new indicator of immune diagnosis of HCV-HCC.
p16, also known as MTS (multiple tumor suppressor 1), is an inhibitor of cyclin dependent kinases such as CDK4 and CDK6. It has been implicated as an important tumor suppressor protein. Missense mutations in p16 gene are strongly linked to several types of human cancer. Deletion of p16 protein will lead to corresponding changes (21,43,44,59). Furthermore, detection of anti-p16 autoantibodies also has been used in early detection of specific changes in HCC. p62/IMP2 is a member of the IGF-II mRNA binding protein (IMP) family and contains four hnRNP K-homology (KH) domains and two RNA recognition motifs (71). Although the sensitivity of anti-p62 autoantibodies is low (44,56,64,72), it is still helpful in the diagnosis of HCC. Cancerous inhibitor of PP2A (CIP2A/p90) is another TAA with molecular weight of 90 kD on SDS-PAGE gel (also known as p90) (73). Current study about the role of p90/CIP2A in tumor formation and progression mainly focus on its ability to inhibit PP2A phosphatase activity and regulate c-myc stability (74). Whether p16, p62 and p90 proteins can induce humoral immune responses in cancer, in other words, whether anti-p16, p62 and p90 autoantibodies can be used as biomarkers for cancer detection still remain to be investigated.
Sui1
Homo sapiens putative translation initiation factor (Sui1), which is also called eukaryotic translation initiation factor1 (eif1), is expressed in some eukaryotic organisms including fungi and yeast (Baker’s yeast). Besides eiF2 and tRNA-Met initiator, Sui1 can directly start the transcription at initiation site. Sui1 is indispensable for discovering the right initiation site and the development of the preinitiation complex (75). The sensitivity, prevalence and specificity of anti-Sui1 autoantibodies for HCC was 10–15.5% (36), 11.7% (9/77) (21) and 98%, respectively.
Binding proteins: Ku86, GPR78, HnRNP L, RalA
The Ku complex is composed of two subunits Ku70 and ku86. It is a DNA dependent protein kinase involving in multiple biological functions. Ku86 is an ATP dependent helicase participating in the repair of DNA rupture. Abnormal expression of Ku86 may lead to the occurrence of some tumors in the digestive system. Xu et al. detected the expression of Ku86 and its autoantibodies in 85 patients with HBV-HCC (40). They further performed immunohistochemistry (IHC) analysis and found that Ku86 was expressed in 67.1% HBV-HCC tissues and HBV-HCC patients with higher expression of Ku86 had a poor prognosis. In addition, the expression of serum anti-Ku86 autoantibody was significantly higher in patients with HCC than in patients with LC and healthy controls (0.47 vs. 0.23 vs. 0.11). Meanwhile, the area under curve (AUC) of detection using anti-Ku86 autoantibodies in combination of AFP reached 0.85, significantly higher than that of using anti-Ku86 autoantibodies alone (40). Nomura et al. detected anti-Ku86 autoantibodies in 57 patients of HCV-HCC and found that the detection sensibility was 60.7% for tumors <2 cm (41). Moreover, the titer of anti-Ku86 autoantibodies differed in patients with HCC at different stages, indicating that it has certain roles in the occurrence and development of HCC.
GPR78, also known as the immune heavy chain binding protein, is a member of HSP70 family originally found in the end of parasitic reticulum. The main function of GPR78 is to help protein folding, collection and transportation. Over-expression of GRP78 was demonstrated on the surface of many cancer cells but not normal cells. Chang et al. found that GPR78 can mediate certain drugs to treat HCC (76). Shao et al. found that the sensibility of anti-GRP78 autoantibodies was 35.5% for serum of HCC patients when using recombinant GRP78 proteins and 71.4% when combined with AFP (47).
HnRNP L is a RNA binding protein that preferentially combines with the repeat sequences of RNA to form hnRNP complexes. Studies have shown that knockout of hnRNP L gene can inhibit cell growth, migration and invasion. Yau et al. used SERPA technology to obtain anti-N segment of hnRNP L autoantibodies from the serum of HBV-HCC patients and showed that its sensitivity was 60% in 70 HCC patients (45).
Ras family small GTP binding protein RalA can be detected in HCC. It has been shown that co-functioning of RalA and RalB play an important role in maintenance of tumorigenicity through regulation of both proliferation and survival. However, the mechanisms of Ral GTPase for proliferation and transformation remain unclear. Wang et al. studied anti-RalA autoantibodies in HCC and found its sensitivity and specificity was 20.1% and 98.3%, respectively (49). Due to the small sample size, whether autoantibodies responding to RalA in HCC sera are correlated with the clinical significance remains to be established.
Survivin
Survivin, a recently described member of the inhibitor-of-apoptosis protein (IAP) family, contains a single baculovirus IAP repeat and lacks a carboxyl-terminal RING finger (77). Yagihashi et al. demonstrated that anti-survivin autoantibodies was present in a substantial fraction of HBV and HCV patients as well as HCC patients at the first time and its sensitivity and specificity were 24.5% and 84.5%, respectively (60). In addition, most researchers considered anti-survivin autoantibodies as one of combined indexes to improve the overall efficiency of diagnosis.
CENPF
Centromere protein F (CENPF) is an essential nuclear protein which was associated with the centromere kinetochore complex. CENPF plays a critical role in chromosome segregation during mitosis (1,27,44). A recent research showed that the sensitivity and specificity of anti-CENPF autoantibodies in HCC were 73.6% and 73.7%, respectively (1). However, Liu et al. showed that the sensitivity and specificity of anti-CENPF autoantibodies were 24.3%, and 100%, respectively (27). This discrepancy suggests that single anti-CENPF autoantibody is not enough for diagnosis of HCC and more things should be done to improve its diagnostic value.
Other autoantibodies in HCC
Other autoantibodies have been explored for diagnosis of HCC including anti-C-myc autoantibodies, anti-HSP60 autoantibodies, anti-HSP70 autoantibodies, anti-NY-ESO-1 autoantibodies, anti-DHCR24 autoantibodies, anti-14-3-3 zeta autoantibodies, anti-nucleophosmin 1 (NPM1) autoantibodies, anti-osteophotin (OPN) autoantibodies and anti-mouse double minute 2 (MDM2) autoantibodies (1,13,15,33,35,38,39,46,78-80). However, most of them have a low sensitivity in diagnosis of HCC and are not explored intensively. The reported sensitivity and specificity of these autoantibodies are below 25% and 70–100%, respectively. A comprehensive analysis of the above various autoantibodies showed that the autoantibodies with low sensitivity and high specificity are suitable biomarkers for diagnosis of HCC. Although autoantibodies are highly specific to HCC at early stage, their low sensitivity limits the application of single autoantibody for detection of HCC. Thus, further exploration is needed to improve their sensitivity. In addition, with the progress of research and the emergence of new experimental techniques, they may play important roles in diagnosis of HCC in the future.
Using the panel of autoantibodies to enhance the sensitivity of HCC diagnosis
Since autoantibodies have low sensitivity in HCC, it is important to use a penal of anti-TAAs autoantibodies to improve the detection rate. Table 2 summarizes some literatures on HCC detection using multiple specific autoantibodies for HCC. Zhang et al. studied single autoantibody and their combinations for detection of HCC (9,56,62,72,81). Zhang et al. showed that the overall sensitivity of eight autoantibodies including anti-p62, anti-Imp1, anti-Koc, anti-p53, anti-c-myc, anti-cyclinB1, anti-p16 and anti-survivin autoantibodies was 59.8% in 142 HCC patients (56). Dai et al. showed that the sensitivity of individual anti-TAAs autoantibodies (anti-CAPERα, anti-p90, anti-RalA, anti-NPM1, anti-MDM2, anti-14-3-3zeta autoantibodies, etc.) was only 6.6–21.1%, while the sensitivity and specificity of their combination were 69.7% and 83%, respectively. This method could make up for the deficiency of AFP detection sensitivity. Moreover, a high sensitivity of the combined autoantibodies was also found in detection of HCC patients at early stage (44).
It is necessary to point out that the sensitivity of combined autoantibodies in detection of HCC varies with the number of autoantibodies (43). Middleton et al. detected 96 cases of HCC patients using 41 autoantibodies and found that although the sensitivity of individual autoantibody for detection of HCC was only 0–10%, the sensitivity and specificity of the combined autoantibodies were 45% and 92%, respectively, significantly different from those in the normal control (NC) group. When the number of combined autoantibodies dropped to 21, the sensitivity only decreased from 45% to 41%, and the specificity only decreased slightly (26). Therefore, the selection of a suitable number of autoantibody in an autoantibodies panel is also one of the most important factors for detection of HCC. Overall, using a panel of autoantibodies in the diagnosis of HCC could achieve significantly higher sensitivity and specificity.
However, excessive autoantibodies in a panel used for detection of HCC in clinical application should be avoided to save resources. Most researchers used less than ten autoantibodies in a panel to detect HCC, which not only improves the detection sensitivity, but also basically meets the diagnostic requirements. Selection of anti-TAAs autoantibodies is the primary key and the most important task in detection of HCC using a panel of autoantibodies.
HCC is heterogeneous and caused by a variety of pathogens (2). A comprehensive analysis and evaluation of various combinations of selected autoantibody-antigen systems will be useful for the development of autoantibody profiles involving different panels or arrays of anti-TAAs autoantibodies, and the results could be helpful for the detection and diagnosis of some types of HCC. Because it is impossible to detect HCC caused by different pathogens through detecting only one abnormal protein, it is necessary to find suitable autoantibodies to establish a panel and improve the detection efficiency of the panel.
Discussion
The establishment of the concept of humoral immunity and immune surveillance of tumor promotes the application of autoantibodies as diagnostic cancer biomarkers. Series of studies on the autoantibodies in HCC have been helpful for the early diagnosis of HCC and enhancing the efficiency of early diagnosis of HCC. Understanding the characteristics of autoantibodies will help us observe the development and progress of disease and predict the prognosis of disease. Many TAAs play important roles in disease development process. Studies on their corresponding autoantibodies can not only provide ideas for clinical diagnosis, but also contribute our understanding on their molecular pathways and signal transduction.
The studies on autoantibodies of HCC are still in the initial exploration stage. There is still lack of a clear conclusion about whether the autoantibodies could be biomarkers of early diagnosis for HCC. For the current studies, autoantibodies have advantage of high specificity in early HCC. In addition, some autoantibodies can also assist the diagnosis of HCC when the expression of AFP is in the normal range. Of course, autoantibodies also have disadvantages. For example, anti-p53 autoantibody is not specific for HCC. Moreover, low sensitivity of autoantibodies also limits their independent application in clinics.
Overall, the development of tumor autoantibodies screening technology has greatly promoted the identification of autoantibodies. Compared with the pathological staging of HCC, serum indices using autoantibodies can predict the occurrence of HCC earlier. Although the studies of autoantibodies in HCC start late and only a few of autoantibodies have been identified to be associated with early HCC, some autoantibodies such as the DCP have been applied in clinics (82). The mechanisms of autoantibodies in HCC need to be further studied. With the development of the screening technology, computer technology and molecular biology technology, the diagnostic value of autoantibodies would further explored and validated and provide more reliable evidences for the diagnosis and prognosis of HCC than before (83).
Acknowledgments
Funding: This work was supported by the Natural Science and Medical Guidance Foundation of Shanghai (grant number 16ZR1400100 and 16411966200) and the National Natural Science Foundation of China, Youth Science Fund Project (grant number 31301187).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.11.70). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hong Y, Long J, Li H, et al. An Analysis of Immunoreactive Signatures in Early Stage Hepatocellular Carcinoma. EBioMedicine 2015;2:438-46. [Crossref] [PubMed]
- Hong Y, Huang J. Autoantibodies against tumor-associated antigens for detection of hepatocellular carcinoma. World J Hepatol 2015;7:1581-5. [Crossref] [PubMed]
- Biselli M, Conti F, Gramenzi A, et al. A new approach to the use of alpha-fetoprotein as surveillance test for hepatocellular carcinoma in patients with cirrhosis. Br J Cancer 2015;112:69-76. [Crossref] [PubMed]
- Liu M, Varela-Ramirez A, Li J, et al. Humoral autoimmune response to nucleophosmin in the immunodiagnosis of hepatocellular carcinoma. Oncol Rep 2015;33:2245-52. [PubMed]
- Desmetz C, Maudelonde T, Mange A, et al. Identifying autoantibody signatures in cancer: a promising challenge. Expert Rev Proteomics 2009;6:377-86. [Crossref] [PubMed]
- Belousov PV, Kuprash DV, Nedospasov SA, et al. Autoantibodies to tumor-associated antigens as cancer biomarkers. Curr Mol Med 2010;10:115-22. [Crossref] [PubMed]
- Tan HT, Low J, Lim SG, et al. Serum autoantibodies as biomarkers for early cancer detection. FEBS J 2009;276:6880-904. [Crossref] [PubMed]
- Dai L, Lei N, Liu M, et al. Autoantibodies to tumor-associated antigens as biomarkers in human hepatocellular carcinoma (HCC). Exp Hematol Oncol 2013;2:15. [Crossref] [PubMed]
- Zhang JY, Tan EM. Autoantibodies to tumor-associated antigens as diagnostic biomarkers in hepatocellular carcinoma and other solid tumors. Expert Rev Mol Diagn 2010;10:321-8. [Crossref] [PubMed]
- Chen X, Fu S, Chen F, et al. Identification of tumor-associated antigens in human hepatocellular carcinoma by autoantibodies. Oncol Rep 2008;20:979-85. [PubMed]
- Chen X, Fu S, Chen F, et al. Identification of tumor-associated antigens in human hepatocellular carcinoma by autoantibodies. Oncol Rep 2008;20:979-85. [PubMed]
- Kuramitsu Y, Nakamura K. Current progress in proteomic study of hepatitis C virus-related human hepatocellular carcinoma. Expert Rev Proteomics 2005;2:589-601. [Crossref] [PubMed]
- Takashima M, Kuramitsu Y, Yokoyama Y, et al. Proteomic analysis of autoantibodies in patients with hepatocellular carcinoma. Proteomics 2006;6:3894-900. [Crossref] [PubMed]
- Heo CK, Woo MK, Yu DY, et al. Identification of autoantibody against fatty acid synthase in hepatocellular carcinoma mouse model and its application to diagnosis of HCC. Int J Oncol 2010;36:1453-9. [PubMed]
- Looi KS, Nakayasu ES, Diaz RA, et al. Using proteomic approach to identify tumor-associated antigens as markers in hepatocellular carcinoma. J Proteome Res 2008;7:4004-12. [Crossref] [PubMed]
- Li SX, Liu LJ, Dong LW, et al. CKAP4 inhibited growth and metastasis of hepatocellular carcinoma through regulating EGFR signaling. Tumour Biol 2014;35:7999-8005. [Crossref] [PubMed]
- Zaenker P, Ziman MR. Serologic autoantibodies as diagnostic cancer biomarkers--a review. Cancer Epidemiol Biomarkers Prev 2013;22:2161-81. [Crossref] [PubMed]
- Wang K, Xu X, Nie Y, et al. Identification of tumor-associated antigens by using SEREX in hepatocellular carcinoma. Cancer Lett 2009;281:144-50. [Crossref] [PubMed]
- Kostianets O, Shyian M, Sergiy D, et al. Serological analysis of SEREX-defined medullary breast carcinoma-associated antigens. Cancer Invest 2012;30:519-27. [Crossref] [PubMed]
- Song MH, Choi KU, Shin DH, et al. Identification of the cancer/testis antigens AKAP3 and CTp11 by SEREX in hepatocellular carcinoma. Oncol Rep 2012;28:1792-8. [PubMed]
- Chen Y, Zhou Y, Qiu S, et al. Autoantibodies to tumor-associated antigens combined with abnormal alpha-fetoprotein enhance immunodiagnosis of hepatocellular carcinoma. Cancer Lett. 2010;289:32-9. [Crossref] [PubMed]
- Zhang Z, Xu L, Wang Z. Screening serum biomarkers for early primary hepatocellular carcinoma using a phage display technique. J Clin Lab Anal 2011;25:402-8. [Crossref] [PubMed]
- Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 1985;228:1315-7. [Crossref] [PubMed]
- Ngubane NA, Gresh L, Ioerger TR, et al. High-throughput sequencing enhanced phage display identifies peptides that bind mycobacteria. PLoS One 2013;8:e77844 [Crossref] [PubMed]
- Talwar H, Rosati R, Li J, et al. Development of a T7 Phage Display Library to Detect Sarcoidosis and Tuberculosis by a Panel of Novel Antigens. EBioMedicine 2015;2:341-50. [Crossref] [PubMed]
- Zhang D, Mao Y, Cao Q, et al. Generation and characterization of a novel recombinant antibody against LMP1-TES1 of Epstein-Barr virus isolated by phage display. Viruses 2013;5:1131-42. [Crossref] [PubMed]
- Liu H, Zhang J, Wang S, et al. Screening of autoantibodies as potential biomarkers for hepatocellular carcinoma by using T7 phase display system. Cancer Epidemiol 2012;36:82-8. [Crossref] [PubMed]
- Gunawardana CG, Memari N, Diamandis EP. Identifying novel autoantibody signatures in ovarian cancer using high-density protein microarrays. Clin Biochem 2009;42:426-9. [Crossref] [PubMed]
- Yang L, Wang J, Li J, et al. Identification of Serum Biomarkers for Gastric Cancer Diagnosis Using a Human Proteome Microarray. Mol Cell Proteomics 2016;15:614-23. [Crossref] [PubMed]
- Jeong JS, Jiang L, Albino E, et al. Rapid identification of monospecific monoclonal antibodies using a human proteome microarray. Mol Cell Proteomics 2012;11:O111.016253.
- Hu CJ, Song G, Huang W, et al. Identification of new autoantigens for primary biliary cirrhosis using human proteome microarrays. Mol Cell Proteomics 2012;11:669-80. [Crossref] [PubMed]
- Roche S, Dauvilliers Y, Tiers L, et al. Autoantibody profiling on high-density protein microarrays for biomarker discovery in the cerebrospinal fluid. J Immunol Methods 2008;338:75-8. [Crossref] [PubMed]
- Oshima Y, Shimada H, Yajima S, et al. NY-ESO-1 autoantibody as a tumor-specific biomarker for esophageal cancer: screening in 1969 patients with various cancers. J Gastroenterol 2016;51:30-4. [Crossref] [PubMed]
- Zhu Q, Han SX, Zhou CY, et al. Autoimmune response to PARP and BRCA1/BRCA2 in cancer. Oncotarget. 2015;6:11575-84. [Crossref] [PubMed]
- Ezzikouri S, Kimura K, Sunagozaka H, et al. Serum DHCR24 Auto-antibody as a new Biomarker for Progression of Hepatitis C. EBioMedicine 2015;2:604-12. [Crossref] [PubMed]
- Zhou JW, Li Y, Yue LX, et al. Autoantibody response to Sui1 and its tissue-specific expression in hepatocellular carcinoma. Tumour Biol 2016;37:2547-53. [Crossref] [PubMed]
- Ying X, Zhao Y, Wang JL, et al. Serum anti-osteopontin autoantibody as a novel diagnostic and prognostic biomarker in patients with hepatocellular carcinoma. Oncol Rep 2014;32:1550-6. [PubMed]
- Liu M, Liu X, Ren P, et al. A cancer-related protein 14-3-3ζ is a potential tumor-associated antigen in immunodiagnosis of hepatocellular carcinoma. Tumour Biol 2014;35:4247-56. [Crossref] [PubMed]
- Liu M, Zheng SJ, Chen Y, et al. Autoantibody response to murine double minute 2 protein in immunodiagnosis of hepatocellular carcinoma. J Immunol Res 2014;2014:906532.
- Xu Y, Liu AJ, Gao YX, et al. Expression of Ku86 and presence of Ku86 antibody as biomarkers of hepatitis B virus related hepatocellular carcinoma. Dig Dis Sci 2014;59:614-22. [Crossref] [PubMed]
- Nomura F, Sogawa K, Noda K, et al. Serum anti-Ku86 is a potential biomarker for early detection of hepatitis C virus-related hepatocellular carcinoma. Biochem Biophys Res Commun 2012;421:837-43. [Crossref] [PubMed]
- EI-Emshaty HM. Serum P53 Abs in HCC patients with viral hepatitis - type C. Hepatogastroenterology 2014;61:1688-95. [PubMed]
- Middleton CH, Irving W, Robertson JF, et al. Serum autoantibody measurement for the detection of hepatocellular carcinoma. PLoS One 2014;9:e103867 [Crossref] [PubMed]
- Dai L, Ren P, Liu M, et al. Using immunomic approach to enhance tumor-associated autoantibody detection in diagnosis of hepatocellular carcinoma. Clin Immunol 2014;152:127-39. [Crossref] [PubMed]
- Yau WY, Shih HC, Tsai MH, et al. Autoantibody recognition of an N-terminal epitope of hnRNP L marks the risk for developing HBV-related hepatocellular carcinoma. J Proteomics 2013;94:346-58. [Crossref] [PubMed]
- Akada J, Kamei S, Ito A, et al. A new type of protein chip to detect hepatocellular carcinoma-related autoimmune antibodies in the sera of hepatitis C virus-positive patients. Proteome Sci 2013;11:33. [Crossref] [PubMed]
- Shao Q, Ren P, Li Y, et al. Autoantibodies against glucose-regulated protein 78 as serological diagnostic biomarkers in hepatocellular carcinoma. Int J Oncol 2012;41:1061-7. [PubMed]
- Liu W, Peng B, Lu Y, et al. Autoantibodies to tumor-associated antigens as biomarkers in cancer immunodiagnosis. Autoimmun Rev 2011;10:331-5. [Crossref] [PubMed]
- Wang K, Chen Y, Liu S, et al. Immunogenicity of Ra1A and its tissue-specific expression in hepatocellular carcinoma. Int J Immunopathol Pharmacol 2009;22:735-43. [PubMed]
- Akere A, Otegbayo JA. Evaluation of the pattern and prognostic implications of anti-p53 in hepatocellular carcinoma. Singapore Med J 2007;48:41-4. [PubMed]
- El Azm AR, Yousef M, Salah R, et al. Serum anti-P53 antibodies and alpha-fetoprotein in patients with non-B non-C hepatocellular carcinoma. Springerplus 2013;2:69. [Crossref] [PubMed]
- Wu M, Mao C, Chen Q, et al. Serum p53 protein and anti-p53 antibodies are associated with increased cancer risk: a case-control study of 569 patients and 879 healthy controls. Mol Biol Rep 2010;37:339-43. [Crossref] [PubMed]
- Gadelhak NA, Gadelhak SA, El-Morsi DA, et al. Prognostic significance of three hepatitis markers (p53 antibodies, vascular endothelial growth factors and alpha fetoprotein) in patients with hepatocellular carcinoma. Hepatogastroenterology 2009;56:1417-24. [PubMed]
- Edis C, Kahler C, Klotz W, et al. A comparison between alpha-fetoprotein and p53 antibodies in the diagnosis of hepatocellular carcinoma. Transplant Proc 1998;30:780-1. [Crossref] [PubMed]
- Kok N, Wiersma IC, Opmeer BC, et al. Sonographic measurement of lower uterine segment thickness to predict uterine rupture during a trial of labor in women with previous Cesarean section: a meta-analysis. Ultrasound Obstet Gynecol 2013;42:132-9. [Crossref] [PubMed]
- Zhang JY, Megliorino R, Peng XX, et al. Antibody detection using tumor-associated antigen mini-array in immunodiagnosing human hepatocellular carcinoma. J Hepatol 2007;46:107-14. [Crossref] [PubMed]
- Li L, Chen SH, Yu CH, et al. Identification of hepatocellular-carcinoma-associated antigens and autoantibodies by serological proteome analysis combined with protein microarray. J Proteome Res 2008;7:611-20. [Crossref] [PubMed]
- Zhou SF, Xie XX, Bin YH, et al. Identification of HCC-22-5 tumor-associated antigen and antibody response in patients. Clin Chim Acta 2006;366:274-80. [Crossref] [PubMed]
- Looi K, Megliorino R, Shi FD, et al. Humoral immune response to p16, a cyclin-dependent kinase inhibitor in human malignancies. Oncol Rep 2006;16:1105-10. [PubMed]
- Yagihashi A, Asanuma K, Kobayashi D, et al. Autoantibodies to survivin in patients with chronic hepatitis and hepatocellular carcinoma. Autoimmunity 2005;38:445-8. [Crossref] [PubMed]
- Lv S, Zhang J, Wu J, et al. Origin and anti-tumor effects of anti-dsDNA autoantibodies in cancer patients and tumor-bearing mice. Immunol Lett 2005;99:217-27. [Crossref] [PubMed]
- Zhang JY. Tumor-associated antigen arrays to enhance antibody detection for cancer diagnosis. Cancer Detect Prev 2004;28:114-8. [Crossref] [PubMed]
- Zhang JY, Zhu W, Imai H, et al. De-novo humoral immune responses to cancer-associated autoantigens during transition from chronic liver disease to hepatocellular carcinoma. Clin Exp Immunol 2001;125:3-9. [Crossref] [PubMed]
- Zhang J, Chan EK. Autoantibodies to IGF-II mRNA binding protein p62 and overexpression of p62 in human hepatocellular carcinoma. Autoimmun Rev 2002;1:146-53. [Crossref] [PubMed]
- Attallah AM, Shiha GE, Ismail H, et al. Expression of p53 protein in liver and sera of patients with liver fibrosis, liver cirrhosis or hepatocellular carcinoma associated with chronic HCV infection. Clin Biochem 2009;42:455-61. [Crossref] [PubMed]
- Atta MM, el-Masry SA, Abdel-Hameed M, et al. Value of serum anti-p53 antibodies as a prognostic factor in Egyptian patients with hepatocellular carcinoma. Clin Biochem 2008;41:1131-9. [Crossref] [PubMed]
- Müller M, Meyer M, Schilling T, et al. Testing for anti-p53 antibodies increases the diagnostic sensitivity of conventional tumor markers. Int J Oncol 2006;29:973-80. [PubMed]
- Parasole R, Izzo F, Perrone F, et al. Prognostic value of serum biological markers in patients with hepatocellular carcinoma. Clin Cancer Res 2001;7:3504-9. [PubMed]
- Shiota G, Kishimoto Y, Suyama A, et al. Prognostic significance of serum anti-p53 antibody in patients with hepatocellular carcinoma. J Hepatol 1997;27:661-8. [Crossref] [PubMed]
- Raedle J, Oremek G, Truschnowitsch M, et al. Clinical evaluation of autoantibodies to p53 protein in patients with chronic liver disease and hepatocellular carcinoma. Eur J Cancer 1998;34:1198-203. [Crossref] [PubMed]
- Nielsen J, Christiansen J, Lykke-Andersen J, et al. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol 1999;19:1262-70. [Crossref] [PubMed]
- Zhang JY, Wang X, Peng XX, et al. Autoantibody responses in Chinese hepatocellular carcinoma. J Clin Immunol 2002;22:98-105. [Crossref] [PubMed]
- Soo Hoo L, Zhang JY, Chan EK. Cloning and characterization of a novel 90 kDa 'companion' auto-antigen of p62 overexpressed in cancer. Oncogene 2002;21:5006-15. [Crossref] [PubMed]
- Junttila MR, Puustinen P, Niemela M, et al. CIP2A inhibits PP2A in human malignancies. Cell 2007;130:51-62. [Crossref] [PubMed]
- Martin-Marcos P, Cheung YN, Hinnebusch AG. Functional elements in initiation factors 1, 1A, and 2beta discriminate against poor AUG context and non-AUG start codons. Mol Cell Biol 2011;31:4814-31. [Crossref] [PubMed]
- Chang YJ, Tai CJ, Kuo LJ, et al. Glucose-regulated protein 78 (GRP78) mediated the efficacy to curcumin treatment on hepatocellular carcinoma. Ann Surg Oncol 2011;18:2395-403. [Crossref] [PubMed]
- Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med 1997;3:917-21. [Crossref] [PubMed]
- Abdou AG, Abd-Elwahed M, Badr M, et al. The Differential Immunohistochemical Expression of p53, c-Jun, c-Myc, and p21 Between HCV-related Hepatocellular Carcinoma With and Without Cirrhosis. Appl Immunohistochem Mol Morphol 2016;24:75-87. [Crossref] [PubMed]
- Zhou SL, Yue WB, Fan ZM, et al. Autoantibody detection to tumor-associated antigens of P53, IMP1, P16, cyclin B1, P62, C-myc, Survivn, and Koc for the screening of high-risk subjects and early detection of esophageal squamous cell carcinoma. Dis Esophagus 2014;27:790-7. [Crossref] [PubMed]
- Megliorino R, Shi FD, Peng XX, et al. Autoimmune response to anti-apoptotic protein survivin and its association with antibodies to p53 and c-myc in cancer detection. Cancer Detect Prev 2005;29:241-8. [Crossref] [PubMed]
- Zhang JY. Mini-array of multiple tumor-associated antigens to enhance autoantibody detection for immunodiagnosis of hepatocellular carcinoma. Autoimmun Rev 2007;6:143-8. [Crossref] [PubMed]
- Takeji S, Hirooka M, Koizumi Y, et al. Des-gamma-carboxy prothrombin identified by P-11 and P-16 antibodies reflects prognosis for patients with hepatocellular carcinoma. J Gastroenterol Hepatol 2013;28:671-7. [Crossref] [PubMed]
- Soussi T. p53 Antibodies in the sera of patients with various types of cancer: a review. Cancer Res 2000;60:1777-88. [PubMed]


