
Regulation of the Mdm2-p53 signaling axis in the DNA damage response and tumorigenesis
The p53 protein and tumor suppression
Proper coordination of p53-responsive gene expression provides a crucial barrier to tumor development, and more than 50% of human cancers harbor mutations in p53 (1,2). Moreover, in cancers bearing wild type p53 alleles, p53 function is frequently compromised through mutations of associated positive regulators or amplification of negative regulators of p53 (1). A large number of p53-mutant mouse models have been developed to study p53 function and regulation. Mice homozygous for p53-null alleles rapidly developed spontaneous tumors (3-5). All p53−/− mice develop tumors by 10 months of age, with a mean time to tumorigenesis of approximately 4.5 months. These tumors are primarily lymphomas (~70–80%, largely T-cell in origin), with some incidence of sarcomas and other tumor types. p53+/− mice develop tumors later than p53−/− mice, with the earliest tumor presentation around 12 months of age (4-6). In contrast, the majority of p53+/− mice (~95%) develop tumors by 24 months of age, with a mean time to tumorigenesis of approximately 17 months. These p53-heterozygous animals present with lymphomas (primarily of B cell origin), osteosarcomas, soft-tissue sarcomas and a range of carcinomas (7). Interestingly, one study observed that while the remaining wild-type p53 allele is deleted in approximately half of p53+/− tumors, the other half retain a functional wild-type allele (8). Thus, a reduction in p53 dosage is alone sufficient to increase the susceptibility of p53+/− cells to tumorigenesis (9).
In agreement with early observations that p53 impaired the ability the ability of oncogenes to transform cells (10,11), numerous studies have shown that combining the p53-null allele with mouse models of cancer, such as those driven by PTEN deficiency, activated BRAFV600E, or Myc overexpression, leads to accelerated tumor development (12-14). Early in vitro studies performed in human cells aimed at identifying the mechanism by which p53 impaired tumor formation identified p53 as a regulator of cell growth, senescence, and apoptosis (15-19). This was supported by subsequent work in mice, wherein p53 was found to regulate spontaneous immortalization of MEFs (20-22), and to be essential for oncogene-induced senescence in cells overexpressing Ras, E2F1 and constitutively active β-catenin (23-25). A role for p53-dependent senescence in tumor suppression in mice was highlighted using mice expressing a mutant p53 protein (p53R172P) that was defective for apoptosis but retaining growth arrest capabilities. These mice displayed delayed Eµ-myc-driven B cell lymphomagenesis compared to mice heterozygous for p53 (26).
The influence of p53 on growth arrest and senescence has been largely attributed to its ability to upregulate p21 expression. p21 protein levels increase in normal fibroblasts as they approach senescence (27) and p21−/− cells display reduced growth arrest in response to DNA damage (28,29). Furthermore, tumor cell lines lacking functional p53 failed to arrest in response to forced expression of p53 when the p21 gene was disrupted (30), and induced expression of p21 promotes senescence in tumor cells lacking functional p53 (31,32).
Studies in primary murine cells have also shown p53 accumulation and stabilization can promote increased apoptosis (33,34), and p53-dependent apoptosis has been shown to suppress Eµ-myc-driven B cell lymphomagenesis (35), E2F1-driven skin carcinomas (36), and brain tumorigenesis induced by a mutant SV40 T antigen (non p53-binding TgT121) (37). Interestingly, development of Eµ-myc-driven tumors is also accelerated in mice lacking the p53-responsive, pro-apoptotic genes Bax (38), or Puma (39-41).
Although p53-dependent growth arrest, senescence and apoptosis appear to be important tumor-suppressive mechanisms, the relative contribution of these mechanisms is likely to be tissue- or cell-type-dependent. This is exemplified by a series of studies in which p53 reactivation in established tumors resulted in apoptosis in lymphomas and senescence in sarcomas, respectively (42-44). More recently, a number of studies have implicated additional p53-dependent mechanisms in impairing tumor growth. Gu and colleagues have described a knock-in mouse in which three p53 acetylation sites in the p53 DNA-binding domain were mutated to arginine (p533KR mice) (45). p533KR mice and cells fail to transactivate the majority of p53 target genes, and induce growth arrest or apoptosis. However, these mice do not develop cancer, suggesting that spontaneous tumor suppression by p53 may occur independently of growth arrest or apoptosis. It is proposed that as these animals retain the ability to transactivate the metabolic targets Gls2 and TIGAR, p53 may display tumor suppressive activity through the regulation of energy metabolism and reactive oxygen species (ROS) levels (45). Similar findings have been reported using, triple knock-out mice deficient for p21, Puma, and Noxa (p21−/−puma−/−noxa−/− mice) (46). These mice are profoundly resistant to DNA damage-associated apoptosis and growth arrest, and largely (but not entirely) resistant to p53-dependent senescence, yet do not develop spontaneous tumors. These authors noted that induction of p53 target genes involved in DNA repair was unperturbed in p21−/−puma−/−noxa−/− mice, and propose that coordination of DNA repair is an essential tumor suppressive activity of p53. More recently, Gu and colleagues have identified the ability of p53 to transcriptionally repress the cysteine/glutamate antiporter SLC7A11 and induce ferroptosis [an iron-dependent mechanism of non-apoptotic cell death (47)] in response to ROS as another mechanism by which p533KR mice may suppress tumorigenesis (48).
The role of p53 responses in the DNA damage response
Following the identification of p53 as a tumor suppressor protein, and in concert with the aforementioned studies elucidating the tumor suppressive activities of p53, a series of studies showed that p53 levels and activity increased in response to DNA damage. Treatment of cells with DNA damaging agents such as ultraviolet light (UV), ionizing radiation (IR), and numerous cancer therapeutic and/or DNA damage-inducing compounds such as diamminedichloroplatinum (cisplatin), mitomycin C, etoposide, hydroxyurea (HU), methyl methanesulfonate (MMS) and actinomycin D results in increased p53 protein levels and associated cell cycle arrest (49-52). Furthermore, p53−/− MEFs are resistant to oncogene-sensitized apoptosis in response to serum withdrawal or a variety of genotoxic agents (53,54). Analyses of p53−/− mice determined that p53 governs IR-induced apoptosis in both thymocytes (55-57) and epithelial stem cells of the small intestine (58). Further studies have identified additional radiosensitive cell populations in the spleen, bone marrow, and hair follicles (59).
An early indication of the signaling pathways governing the p53 response to DNA damage came from analysis of from patients with ataxia-telangiectasia; an autosomal recessive disorder resulting in neuronal degeneration, sensitivity to IR, premature ageing, increased incidence of cancer and other pathologies. Patients’ cells do not display increased p53 levels and activity following IR exposure (60), and mice null for ataxia telangiectasia mutated (ATM) are extremely sensitive to IR-induced lethality, and show profound defects in DNA damage-induced growth arrest and apoptosis (61-64). Similar to AT patient cells, p53 is not stabilized in ATM−/− MEFs or thymocytes following IR (63,65). Furthermore, these animals succumb to T-cell lymphomas by 6 months of age (61,62). Research aimed at identifying the mechanism by which ATM leads to p53 stabilization is discussed below.
Regulation of p53 function
The chief negative regulator of p53 stabilization and activity is the Mdm2 oncoprotein. The murine double minute 2 (Mdm2) gene was initially identified as an amplified DNA sequence present in a spontaneously immortalized mouse 3T3 cell line (66). Mdm2 overexpression is capable of conferring tumorigenicity (67), and Mdm2 is amplified in a significant fraction (~30%) of soft tissue sarcomas (68-70). Further studies have identified Mdm2 amplification in a variety of other tumor types, including breast carcinomas (71), glioblastomas and astrocytomas (72), myeloid neoplasms (73), B cell lymphomas (74) and oral carcinomas (75).
Shortly after the identification of Mdm2’s interaction with p53, mapping of the p53 and Mdm2 interaction domains determined that the N-terminus of Mdm2 bound to and inhibited the transactivation domain of p53 (76,77). Accordingly, Mdm2 overexpression cooperates with Ras in transforming primary cells (78), and inhibits p53-dependent growth arrest and apoptosis in various cell lines (79-81).
It was subsequently shown that Mdm2 can also promote the proteasomal degradation of p53 (82,83) by functioning as an E3 ubiquitin ligase and directing p53 polyubiquitination (84). This E3 activity of Mdm2 is dependent on its C-terminal RING finger domain (85), which also promotes the nuclear export of p53 by directing its monoubiquitination (86-89). It is proposed that low levels of Mdm2 activity induce monoubiquitination and nuclear export of p53, whereas high levels promote polyubiquitination and nuclear degradation of p53 (90). The principle sites on p53 of Mdm2-ubiquitin ligation are a series of C-terminal lysines also targeted by acetylation, creating one of many layers of regulation of p53 stability and activity (91-94). Notably, the Mdm2 gene is also a target of the p53 transcription factor (95,96). As p53 becomes stabilized and active, it increases the expression levels of its own negative regulator Mdm2, forming an autoregulatory feedback loop that returns p53 protein and activity to basal levels. Furthermore, Mdm2 has also been shown to be capable of directing its own degradation (85,97).
The crucial role of Mdm2 in regulating p53 activity is illustrated by the p53-dependent lethality of Mdm2-null mice during early embryogenesis. Mdm2−/− mice display peri-implantation lethality due to unregulated p53 activity, and crossing these mice with the p53-null allele completely rescues Mdm2-null mice (98,99). Mice null for both Mdm2 and p53 develop spontaneous tumors of similar incidence and spectrum as p53-null mice (100). Furthermore, primary p53-null and Mdm2/p53 double-null cells display similar growth characteristics in culture and in their response to genotoxic agents (100). Mdm2+/− mice display delayed Myc-driven lymphomagenesis (101), and hypomorphic-Mdm2 mice expressing reduced levels of Mdm2 displayed p53-dependent sensitivity to radiation-induced lethality and apoptosis in lymphopoietic tissues (102). Thus, perturbations in the levels of Mdm2 can significantly impact p53 responses to oncogenes and DNA damage.
In contrast with Mdm2-null mice, mice overexpressing an Mdm2 transgene are viable and succumb to spontaneous tumors (103). The rate of tumorigenesis in Mdm2-transgenic mice is slower than that observed in p53−/− or p53+/− mice, likely due to the relatively modest levels of Mdm2 overexpression (approximately 4 fold) in the Mdm2-transgenics. However, like p53-mutant mice, Mdm2-transgenics present with a large percentage of lymphomas and sarcomas. Notably, while presence of the Mdm2-transgene does not accelerate spontaneous tumorigenesis in p53−/− mice, it does increase their incidence of sarcomas, revealing a p53-independent contribution of Mdm2 overexpression to tumorigenesis. Mdm2 overexpressing mice also display accelerated Myc-driven lymphomagenesis (104). This same study showed that elevated levels of Mdm2 resulted in reduced p53 protein levels and activity in B cells, and reduced B cell apoptosis following IR.
Similar to Mdm2, the related protein MdmX (or Mdm4) is also capable of binding p53 and inhibiting p53 transactivation of target genes (105,106). MdmX and Mdm2 share 34% protein homology and contain highly homologous p53-binding, acidic, zinc finger, and RING finger domains (105). As with Mdm2, the MdmX gene is amplified or overexpressed in a variety of tumor types (107-111). However, unlike Mdm2, MdmX does not possess the ability to directly ubiquitinate p53 (112,113).
Mice null for MdmX display a similar p53-dependent embryonic lethality as observed in Mdm2−/− mice, albeit later in development (E9.5–10.5), and are rescued by deletion of p53 (114-116). Notably, the lethality in MdmX−/− embryos appears to be predominantly associated with a lack of proliferation, as opposed to aberrant apoptosis [as reported for Mdm2−/− embryos (117)]. Accordingly, a subsequent study revealed that co-deletion of p21 can significantly delay the embryonic lethality of MdmX-null mice (118). MdmX+/− cells and mice display decreased oncogene-driven transformation and Eµ-myc-driven lymphomagenesis, respectively (119), and MdmX+/− mice are sensitized to radiation induced lethality (119).
Mdm2 and MdmX have been shown to interact via their C-terminal RING domains (120,121), and stabilization of Mdm2 by heterodimerization with MdmX increases the ability of Mdm2 to degrade p53Jeny (122-125). Notably, the turnover of MdmX is mediated by Mdm2Jeny (126-128).
Recently, a series of Mdm2 and MdmX knock-in mouse models have been generated that display altered Mdm2-MdmX interactions and/or Mdm2 E3 ligase activity (129-132). Analyses of these models have revealed that Mdm2-MdmX interactions are crucial for inhibiting p53 activity during development and tissue homeostasis, whereas the E3 ligase function of Mdm2 is vital for regulating p53 protein levels and activity in cellular and organismal responses to DNA damage (132).
p53 stabilization and activation in response to cell stress
The inhibitory role of MDM proteins on p53 protein stabilization and activities must be interrupted in order for p53 to become elevated and activated in response to DNA damage or other forms of stress.
An important mediator of oncogene-dependent activation of p53 is the tumor suppressor protein p19ARF (p14ARF in humans). Oncogenes including c-Myc, Ras and E1A induce ARF and cause p53-dependent growth arrest and apoptosis (133-135). ARF binds to Mdm2 and can block its ubiquitin ligase activity towards p53Jeny (136-141) as well as sequester Mdm2 in the nucleolus (142,143). This facilitates p53 protein stabilization and activation in order to limit the transformative effects of aberrant oncogene activity.
DNA damage-induced modifications of p53
The cellular response to DNA damage is primarily governed by the PI3K-related serine/threonine kinases (PIKKs) ATM and ataxia telangiectasia and Rad3-related protein (ATR). ATM is activated by DNA damaging agents that create DSBs, while ATR is activated following recruitment to ssDNA regions. The related protein DNA-PK (DNA-dependent protein kinase) primarily regulates a smaller group of proteins involved in DSB end joining. Following the recognition of DNA damage by different sensor proteins, these kinases trigger the direct or indirect phosphorylation of numerous effector proteins involved in a multitude of signaling networks that promote different DNA repair processes, cell-cycle arrest and programmed cell death (144,145).
Included among the various PIKK substrates is p53. p53 is phosphorylated on a number of residues, primarily clustered in the N- and C-terminal regions (Figure 1), in response to various DNA-damaging agents (146,147). Phosphorylation of C-terminal residues is primarily thought to influence site-specific DNA binding by p53, whereas N-terminal phosphorylation events have been implicated in regulating the Mdm2-p53 interaction as well as p300/CBP recruitment (146,147). Seven serines (Ser6, 9, 15, 20, 33, 37, 46) and two threonines (Thr18 and 81) in the N-terminal region of human p53 are phosphorylated in response to exposing cells to IR or UV light (146). The majority of these phosphorylation events occur directly by ATM, ATR, or DNA-PK (148-151) or indirectly by the ATR- and ATM-activated checkpoint kinases Chk1 or Chk2Jeny (152-154). Additionally, casein kinase 1 (CK1) has also been shown to phosphorylate a number of these residues (155,156).
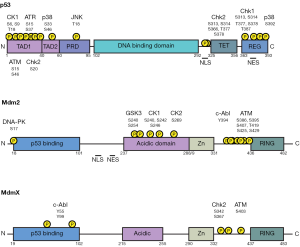
Of particular interest in the search for the mechanism of p53 stabilization following DNA damage were residues Ser15 and Ser20 (Ser18 and Ser23 in mice). In vitro experiments revealed phosphorylation of Ser15, a target of both ATM and ATR, inhibits the p53-Mdm2 interaction (157) and coincides with p53 activation (158). Similar experiments have shown phosphorylation of Ser20, a target of Chk2, leads to reduced Mdm2-medated degradation of p53 and increased p53 activity (153,159,160). Furthermore, Thr18 phosphorylation, which occurs through CK1 and can disrupt Mdm2-p53 binding, was shown to be dependent on prior Ser15 phosphorylation (155,161).
As these p53 residues are located within, or immediately adjacent to, the Mdm2-p53 binding interface (162), it was hypothesized that phosphorylation of these residues was sufficient to account for p53 stabilization and activation following DNA damage. However, analysis of various genetically engineered mouse models, which allowed for the examination of these phosphorylation events under endogenous conditions, revealed that these phosphorylation events were insufficient to account for the full effects of DNA damage on p53 stabilization and activation or for p53 tumor suppression.
p53S18A mice in which serine 18 (Ser15 in humans) is replaced with alanine show no significant defects in p53 protein stabilization in thymocytes or MEFs in response to DNA damage (163,164). MEFs from p53S18A mice show no defects in proliferation or growth arrest after DNA damage, while thymocytes show an intermediate (compared to p53−/−) defect in apoptosis. However, the ability of p53 to transactivate a number of target genes is compromised in p53S18A mice. A recent report suggests that this may be due to a role for p53 Ser15 in transcription and promoter relaxation as opposed to p53 stabilization (165). Additionally, p53S18A mice develop Eµ-myc-driven B cell lymphomas at an accelerated rate, possibly due to their apoptotic defects (166).
p53S23A mice in which serine 23 (Ser20 in humans) is replaced with alanine, show a similar absence of defects in p53 stabilization or growth arrest MEFs (167,168). However, p53S23A mice do show reduced stabilization of p53 and apoptosis in thymocytes in response to IR, though intermediate compared to p53−/− thymocytes (168). Furthermore, p53S23A mice develop spontaneous tumors (predominantly B cell lymphomas) beginning at approximately 12 months, with 70% of animals having developed tumors by 24 months. Interestingly, p53S18A/S23A mice, in which both Ser18 and Ser23 have been substituted, display more profound deficiencies in p53 stabilization and function, indicating an additive effect of phosphorylation of these two residues in regulating p53 function (169). While still intermediate to the phenotypes observed in ATM−/− and p53−/− cells, thymocytes from p53S18A/S23A mice show more significantly impaired p53 stabilization, transactivation of target genes, and apoptosis in response to irradiation than either single-mutant alone. However, p53 stabilization and activities are still unperturbed in MEFs. These mice are similarly tumor prone as reported for Ser23 mutant mice, and again present primarily with lymphomas.
DNA damage-induced modifications of MDM proteins
As the in vivo results obtained from p53 knock-in mice failed to replicate the profound defects in DNA damage-induced p53 stabilization and activity predicted by in vitro studies, additional signaling events must contribute to this process. In addition to p53, its primary negative regulators Mdm2 and MdmX are also subject to a multitude of phosphorylation events in response to DNA damage (Figure 1).
Phosphorylation of MDM proteins by ATM
In response to DNA damage, human MdmX is phosphorylated at Ser342 and Ser367 by Chk2Jeny (170-173) and Ser403 by ATM (174). These phosphorylation events lead to MdmX degradation, concurrent with p53 stabilization and activation (170,171). This phosphorylation-dependent degradation of MdmX is proposed to be directed by Mdm2, and possibly mediated by changes in MdmX binding by 14-3-3 and the deubiquitinase HAUSP (171-173,175). Wahl and colleagues have generated an MdmX3SA mouse model in which all three of these serine residues are replaced with alanine (176). MdmX3SA mice display impaired p53 stabilization and decreased p53 activity in response to IR. Furthermore, MdmX3SA mice are resistant to lethal doses of IR, and though not prone to spontaneous tumorigenesis, these mice display increased Eµ-myc-driven lymphomagenesis. Thus, DNA damage-induced phosphorylation of MdmX, a negative regulator of p53, can impact p53 stabilization and activity.
ATM-dependent phosphorylation of Mdm2 also precedes p53 stabilization after DNA damage (177). It was initially shown that ATM directly phosphorylates Ser395 of human Mdm2 (Ser394 in mouse) in response to DNA damage (178). Furthermore, in transfection-based assays, Mdm2 with an aspartic acid in place of Ser395 (mimicking phosphorylation) shows a decreased capacity to induce p53 degradation and nuclear export (178). The phosphatase Wild-type p53-induced phosphatase 1 (Wip1) can dephosphorylate Mdm2 Ser395, and dephosphorylated Mdm2 has increased stability and affinity for p53, facilitating p53 ubiquitination and degradation (179). This result trends with another study that showed that DNA damage-induced p53 stabilization is preceded by the destabilization of Mdm2, which is a phenomenon that could be inhibited with the PIKK-inhibitor wortmannin (180). However, a subsequent study identified five additional residues in the C-terminal region of human Mdm2 that are phosphorylated by ATM (Ser386, Ser407, Thr419, Ser425 and Ser 429) (181) and in vitro work in which all six residues were replaced with alanine or aspartic acid suggests that ATM phosphorylation of this series of residues inhibits RING domain oligomerization and E3 ligase activity (181,182). Thus, ATM phosphorylation of Mdm2 is proposed to influence both Mdm2 stability and activity towards p53.
In order to examine the impact of Mdm2 Ser395 phosphorylation under endogenous conditions, our lab has previously reported the generation and initial characterization of a mouse model wherein ATM phosphorylation of Mdm2 at serine residue 394 (the equivalent of human Mdm2 Ser395) was abolished (Mdm2S394A mice) (183). Cells and tissues in Mdm2S394A mice display profound defects in DNA damage-induced p53 protein stabilization and p53 target gene activation. This failure to induce a robust p53 response translates to less p53-dependent apoptosis in hematopoietic tissues, radio-resistance, and increased spontaneous lymphomagenesis. Furthermore, replacing Mdm2 Ser394 with aspartic acid (Mdm2S394D mice) and mimicking constitutive phosphorylation at this residue result in prolonged p53 activity following damage and suggest dephosphorylation of this residue is involved in resolving the p53 response (183). Therefore, ATM phosphorylation of the negative regulator of p53, Mdm2, can profoundly impact p53 stabilization and activation in response to stress in vivo. In a subsequent study using Mdm2S394A mice we showed that phosphorylation of Mdm2-S394 regulates p53 activity and the DNA damage response in lymphatic tissues in vivo by modulating Mdm2 stability (184). Intriguingly, while Mdm2-S394 phosphorylation delays lymphomagenesis in Eµ-myc transgenic mice, and preventing Mdm2-S394 phosphorylation obviates the need for p53 mutation in Myc-driven tumorigenesis, irradiated Mdm2S394A mice display increased hematopoietic stem and progenitor cell functions, and decreased radiation-induced lymphomagenesis. These findings document contrasting effects of ATM-Mdm2 signaling on p53 tumor suppression (184).
Phosphorylation of MDM proteins by c-Abl
Similar to ATM, the c-Abl tyrosine kinase is activated by a variety of DNA damaging agents (185-187). c-Abl interacts with ATM and is phosphorylated on Ser465, leading to its activation (188,189). This initially led to c-Abl activities in the DNA damage response being viewed as downstream of ATM. However, more recent work has shown c-Abl to phosphorylate both ATM and ATR, and that these phosphorylation events are required for maximal activity of either PIKK (187). Overexpression studies indicate c-Abl promotes growth arrest in a p53-dependent manner, and apoptosis by p53-dependent and independent mechanisms (190-193). c-Abl mediated p53-independent apoptosis is attributed to the p53 homolog p73, which is directly phosphorylated by c-Abl on Tyr99Jeny (194-196). However, no c-Abl target residues have been identified on p53.
MdmX is phosphorylated by c-Abl on Tyr55 and Tyr99 in response to DNA damage. These residues are located within the p53 binding domain of MdmX, and Tyr99 phosphorylation impairs p53 binding in a transfection-based assay (197). Similar co-expression studies have shown that c-Abl protects p53 from Mdm2 mediated degradation, and overcomes the inhibitory effect of Mdm2 on p53 transcriptional activity and p53-dependent apoptosis (198). Additionally, c-Abl is required for maximal p53 accumulation in response to IR, doxorubicin, and mitomycin C in MEFs, and co-expression of c-Abl overcomes Mdm2 mediated ubiquitination and nuclear export of p53 (199). In vitro studies have shown that c-Abl phosphorylates human Mdm2 on Tyr394 (Tyr393 in mouse) as well as Tyr276 and Tyr405 (200,201), and that c-Abl phosphorylation of Mdm2 Tyr394 impairs Mdm2’s ability to inhibit p53’s stabilization and transactivation, and p53-mediated apoptosis (200). It has since been proposed that c-Abl phosphorylation of Mdm2 increases Mdm2-MdmX binding and promotes Mdm2-directed MdmX ubiquitination. This increase in MdmX ubiquitination ultimately destabilizes the Mdm2-MdmX complex, promoting p53 stabilization (202). Recently, our lab has very recently generated Mdm2-Tyr393 knock-in mice to explore the physiological role of c-Abl phosphorylation of Mdm2 on p53 stabilization and activation. However, analysis of the DNA damage response and tumorigenesis in these mice is presently ongoing.
Other phosphorylation events on Mdm2
Located in the acidic domain of Mdm2 is a cluster of residues that are phosphorylated under homoeostatic conditions (203). Phosphorylation of these residues is known to occur through the activities of the kinases—glycogen synthase kinase 3 beta (GSK-3β), CK1, and CK2Jeny (204-207). Phosphorylation of these residues improves Mdm2-mediated turnover of p53 in the absence of stress stimuli (204,207,208), and hypo-phosphorylation of this region of Mdm2 is reported to coincide with DNA damage-induced p53 stabilization (208). Accordingly, inhibition of GSK-3β leads to p53 stabilization in cells (207). Notably, GSK-3β is inhibited through phosphorylation by Akt, which is activated by DNA-PK following DNA damage (209). Akt-mediated inhibition of Mdm2 activity in this context contrasts with other reports in which the direct phosphorylation of Mdm2 Ser166 and Ser186 by Akt is proposed to inhibit p53 activity by facilitating Mdm2 translocation into the nucleus (210,211) and by inhibiting Mdm2 self-ubiquitination and degradation (212).
Effects of Mdm2 phosphorylation on p53 function
Previous studies have suggested that ATM phosphorylation of the analogous residue on human MDM2 (Ser395), either alone or in combination with several other ATM-target serine residues in the same region, impacts the ability of Mdm2 to promote p53 degradation and nuclear export, and governs Mdm2 RING-domain oligomerization and polyubiquitination of p53 (178,181). It is likely that DNA damage-induced p53 activity is caused not only by reduced Mdm2-mediated p53 degradation (due to destabilization of Mdm2) but also by reduced Mdm2 steric inhibition of p53. As Mdm2 binds to the amino-terminal, transcriptional activation domain of p53 and inhibits p53 target gene expression, reduced Mdm2-p53 complex formation after Mdm2 phosphorylation by ATM may account for an increase in p53 activity even when p53 protein stability is only modestly altered (76,213).
As discussed above, we and others have demonstrated that Mdm2 phosphorylation can reduce Mdm2 stability, but it remains unclear how Mdm2 phosphorylation facilitates Mdm2 destabilization. One study has suggested that Mdm2 stability is primarily mediated through self-ubiquitination (180). This study reached this conclusion by overexpressing an Mdm2 mutant with abrogated RING E3 activity (Mdm2C464A) in U2OS cells. However, a subsequent study in which MEFs from the corresponding Mdm2C462A knock-in mice were examined showed that Mdm2 RING E3 activity was in fact dispensable for Mdm2 destabilization after IR (129).
Degradation of Mdm2 in the absence of Mdm2 ubiquitin ligase function is indicative of other ubiquitin ligases being capable of regulating Mdm2 stability. One study has shown that the p300/CBP-associated factor (PCAF) is capable of promoting Mdm2 ubiquitination, and that PCAF can impact Mdm2 levels under unstressed conditions as well as Mdm2 destabilization in response to DNA damage (214). Similarly, the anaphase-promoting complex/cyclosome (APC/C) E3 ubiquitin ligase complex has been shown to ubiquitinate Mdm2, with siRNA-mediated depletion of APC2, the Mdm2-binding member of the APC/C complex, leading to Mdm2 accumulation and diminished p53 stabilization in response to DNA damage (215). Other studies have identified the F-box proteins β-TRCP and FBXO31 as mediators of Mdm2 degradation by the SCF complex (216,217). β-TRCP mediated degradation of Mdm2 is dependent on Mdm2 phosphorylation by CK1 and promotes Mdm2 turnover in response to DNA damage (216). This is seemingly in contradiction with previous studies showing hypo-phosphorylation of CK1 target residues on Mdm2 following DNA damage (208). Notably, knockdown of β-TRCP does not appear to impact the initial DNA damage-induced stabilization of p53, but rather Mdm2 stability and p53 levels at later time points in the damage response (216). It has subsequently been shown that ATM phosphorylates CK1, and that this promotes CK1 nuclear localization and Mdm2 degradation (218). It is conceivable that the phosphorylation state of Mdm2 at CK1 target residues has opposing effects on Mdm2 stability depending on the damage response phase and differing active signaling events. Contrastingly, FBXO31 knockdown in cell lines abolishes the initial Mdm2 destabilization and p53 stabilization following DNA damage (217). FBXO31 can direct the polyubiquitination and degradation of Mdm2, and the FBXO31-Mdm2 interaction appears dependent on ATM phosphorylation of Mdm2 (217). However, the requirement for ATM phosphorylation was determined through the expression of an Mdm2 protein lacking all 6 proposed ATM target residues, and conversely, the treatment of lysates with phosphatase or cells with an ATM kinase inhibitor. Consequently, the precise mechanism of how ATM phosphorylation of Mdm2 Ser394 specifically promotes Mdm2 degradation under endogenous conditions remains to be determined.
In opposition to Mdm2 destabilization, a number of different mechanisms are proposed to promote Mdm2 stability. These mechanisms include modifications of Mdm2 with the small, ubiquitin-like proteins SUMO (Small ubiquitin-related modifier) and NEDD8 (Neural precursor cell expressed developmentally down-regulated protein 8), which are separately proposed to have Mdm2 stabilizing effects and to decrease in response to DNA damage (219,220). Additionally, Mdm2, MdmX and p53 can be deubiquitinated by the HAUSP (herpesvirus-associated ubiquitin-specific protease) protein (175,221,222). DNA damage has been shown to reduce the affinity of Mdm2 and MdmX for HAUSP, leading to their enhanced ubiquitination (175). A proposed mechanism for the dissociation of Mdm2 from HAUSP involves ATM phosphorylation of Daxx (death domain-associated protein 6) triggering its dissociation from Mdm2, and relieving Daxx mediated promotion of Mdm2-HAUSP interaction (223,224). MDM2 is also been reported to be deubiquitinated by the ubiquitin-specific proteases USP2a and USP15 (225,226). Notably, USP2a also acts as a deubiquitinating enzyme for MdmX, and is downregulated in response to cisplatin (227).
Therapeutic implications
That the overwhelming majority of tumors display loss of p53 function has led to massive efforts to develop cancer therapies that either exploits p53 tumor suppressive function, or its propensity for mutation or inactivation. These efforts have included, but are not limited to, the development of gene therapies involving viral delivery of p53 expression vectors into tumors (228), therapies employing siRNA-mediated knockdown of negative regulators of p53 such as viral E6 protein (229) and Mdm2 (230), and immunotherapies targeting elevated levels of p53 protein observed following p53 mutation (231).
As extensive research has provided an ever-improving understanding of the structures and activities of the various p53-interacting proteins involved in the p53 signaling network, a host of small-molecule and peptide therapeutics have also been identified that facilitate the pharmaceutical control of p53 signaling. Approximately 50% of tumors retain expression of wild-type p53. Accordingly, these tumors are potential targets for therapies that stimulate the tumor suppressive activities of p53 by freeing it from negative regulation. An obvious target for this approach is the interaction of p53 with its primary negative regulator, Mdm2. Aided by the compact, “druggable” p53-Mdm2 interface, which involves a short helical fragment of p53 binding in a deep hydrophobic pocket of Mdm2 (162), a series of small-molecule inhibitors of the p53-Mdm2 interaction have been developed. Both high throughput screening and structure based design methods have yielded three classes of small-molecule inhibitors, the Nutlins (232), the benzodiazepinediones (233), and the spiro-oxindoles (234). By interfering with p53-Mdm2 binding, these compounds have been shown to activate p53 function, and optimized derivatives of these compounds are currently in a number of clinical trials (235). However, these compounds do not strongly inhibit the MdmX-p53 interaction, due to differences in the p53 binding pocket of MdmX as compared to Mdm2, and do not always show effects on cancer cells expressing high levels of MdmX (236,237). More recently, several small-molecules that target the p53-MdmX interaction have been reported (WK 298 and SJ-172550) (238,239), as well as molecules that inhibit MdmX transcription (NSC 207895) (240) or stability (17-AAG) (241). Furthermore, compounds that inhibit both p53-Mdm2 and p53-MdmX binding have been generated and are being studied (RO-5963) (242). Finally, the small-molecule RITA (reactivation of p53 and induction of tumor cell apoptosis) was identified in a high-throughput screen as inducing p53 accumulation and activation by binding p53 itself. RITA binding is proposed to induce a conformational change in p53, resulting in its dissociation from Mdm2 (243). However, it has also been shown that RITA causes DNA-protein crosslinks and is metabolized to a reactive species, leading to p53-independent toxicity (244-246).
A separate class of inhibitors of p53-Mdm2 and p53-MdmX binding are the stabilized small peptides termed “stapled peptides” (247,248). These compounds employ a chemical strategy termed “hydrocarbon stapling” that fixes an all-hydrocarbon crosslink within synthetic peptides to preserve their α-helical structure, confer protease resistance, and promote their cellular uptake (249). One of these compounds, SAH-p53-8 is capable of binding to both Mdm2 and MdmX with high affinity (247,248). However, despite a higher affinity for Mdm2 than Nutlin 3a, SAH-p53-8 is less potent at disrupting the p53-Mdm2 interaction in cells, possibly due to prevailing issues with bioavailability (250).
Compounds have also been developed that inhibit the ubiquitin ligase function of Mdm2, such as HLI98 compounds (251) and the related MPD compounds (252), and MEL23 and MEL24 (253). These compounds stabilize p53 and Mdm2 promote p53 transcriptional activation and growth arrest and apoptosis. However, these inhibitors also elicit some p53—independent cytotoxicity, particularly at higher concentrations, possibly due to inhibition of other RING domain-containing E3 ligases.
Separate from therapies aimed at combating tumor growth through p53 induction, research has also focused on regulating the p53 response in non-malignant tissues, in an effort to reduce the side effects of cytotoxic cancer therapies such as cytopenia and hair loss. In one approach, using the concept of cyclotherapy, an initial nongenotoxic p53 inducing compound is used to promote a reversible cell cycle arrest in normal proliferative tissues, before a second drug is employed to kill proliferating cells, presumably only the p53 mutant tumor cells. This approach has shown promise in cell lines, using Nutlin compounds for p53 induction (254,255). However, this approach is specific for chemotherapeutics that target S or M phase, such as nucleoside analogs β-D-arabinofuranoside (Ara-C) and gemcitabine, or paclitaxel, and does not show effectiveness when Nutlins are used in combination with doxorubicin or cisplatin. Conversely, the compound Pifithrin-α (PFT-α, an abbreviation for “p-fifty-three inhibitor”), a p53-inhibiting compound identified in a high-throughput screen, has been shown to protect p53 wild-type cells from apoptosis induced by irradiation and the cytotoxic drugs doxorubicin, etoposide, Taxol, and Ara-C (256).
In another approach that exploits the propensity for p53 inactivation in tumor cells, Yuan and colleagues have identified a phenomenon in which pre-treatment of cells with low-dose arsenic induces p53 stabilization and a p53-dependent metabolic shift from oxidative phosphorylation to anaerobic glycolysis which confers protection to normal tissues from 5FU and radiation-induced toxicities (257-259). This approach has recently shown hematopoietic protection in humans receiving myelosuppressive chemotherapy in a recently reported clinical trial (260).
Our recent findings suggest an avenue for an additional layer of pharmaceutical control of the p53 pathway. We have shown Mdm2 phosphorylation to significantly impact the capacity to repopulate bone marrow following irradiation and (in the case of Ser394 phosphorylation) simultaneously protect against lymphomagenesis induced by repeated IR exposure (184). Similar effects have been observed in the absence of appropriate MdmX phosphorylation (176). While broad inhibition of DNA damage responsive kinases such as ATM, Chk2 and c-Abl would likely be undesirable due to their involvement in additional processes such as DNA repair, small-molecule therapeutics that inhibit DNA damage-induced Mdm2 or MdmX phosphorylation events may be useful in reducing unwanted chemotherapeutic side effects without compromising p53 tumor suppressive function.
Acknowledgments
Funding: This work was supported by the National Institutes of Health (CA077735 to SN Jones).
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Zhi-Min Yuan) for the series “p53 Biology and Cancer” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.11.75). The series “p53 Biology and Cancer” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell 1997;88:323-31. [Crossref] [PubMed]
- Soussi T, Beroud C. Assessing TP53 status in human tumours to evaluate clinical outcome. Nat Rev Cancer 2001;1:233-40. [Crossref] [PubMed]
- Donehower LA, Harvey M, Slagle BL, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 1992;356:215-21. [Crossref] [PubMed]
- Purdie CA, Harrison DJ, Peter A, et al. Tumour incidence, spectrum and ploidy in mice with a large deletion in the p53 gene. Oncogene 1994;9:603-9. [PubMed]
- Jacks T, Remington L, Williams BO, et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol 1994;4:1-7. [Crossref] [PubMed]
- Harvey M, McArthur MJ, Montgomery CA Jr, et al. Spontaneous and carcinogen-induced tumorigenesis in p53-deficient mice. Nat Genet 1993;5:225-9. [Crossref] [PubMed]
- Donehower LA, Lozano G. 20 years studying p53 functions in genetically engineered mice. Nat Rev Cancer 2009;9:831-41. [Crossref] [PubMed]
- Venkatachalam S, Shi YP, Jones SN, et al. Retention of wild-type p53 in tumors from p53 heterozygous mice: reduction of p53 dosage can promote cancer formation. EMBO J 1998;17:4657-67. [Crossref] [PubMed]
- Venkatachalam S, Tyner SD, Pickering CR, et al. Is p53 haploinsufficient for tumor suppression? Implications for the p53+/- mouse model in carcinogenicity testing. Toxicol Pathol 2001;29:147-54. [Crossref] [PubMed]
- Finlay CA, Hinds PW, Levine AJ. The p53 proto-oncogene can act as a suppressor of transformation. Cell 1989;57:1083-93. [Crossref] [PubMed]
- Eliyahu D, Raz A, Gruss P, et al. Participation of p53 cellular tumour antigen in transformation of normal embryonic cells. Nature 1984;312:646-9. [Crossref] [PubMed]
- Chen Z, Trotman LC, Shaffer D, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 2005;436:725-30. [Crossref] [PubMed]
- Dankort D, Filenova E, Collado M, et al. A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes Dev 2007;21:379-84. [Crossref] [PubMed]
- Eischen CM, Weber JD, Roussel MF, et al. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev 1999;13:2658-69. [Crossref] [PubMed]
- Baker SJ, Markowitz S, Fearon E, et al. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science 1990;249:912-5. [Crossref] [PubMed]
- Diller L, Kassel J, Nelson CE, et al. p53 functions as a cell cycle control protein in osteosarcomas. Mol Cell Biol 1990;10:5772-81. [Crossref] [PubMed]
- Chen PL, Chen Y, Bookstein R, et al. Genetic mechanisms of tumor suppression by the human p53 gene. Science 1990;250:1576-80. [Crossref] [PubMed]
- Michalovitz D, Halevy O, Oren M. Conditional inhibition of transformation and of cell proliferation by a temperature-sensitive mutant of p53. Cell 1990;62:671-80. [Crossref] [PubMed]
- Martinez J, Georgoff I, Martinez J, et al. Cellular localization and cell cycle regulation by a temperature-sensitive p53 protein. Genes Dev 1991;5:151-9. [Crossref] [PubMed]
- Harvey DM, Levine AJ. p53 alteration is a common event in the spontaneous immortalization of primary BALB/c murine embryo fibroblasts. Genes Dev 1991;5:2375-85. [Crossref] [PubMed]
- Harvey M, Sands AT, Weiss RS, et al. In vitro growth characteristics of embryo fibroblasts isolated from p53-deficient mice. Oncogene 1993;8:2457-67. [PubMed]
- Tsukada T, Tomooka Y, Takai S, et al. Enhanced proliferative potential in culture of cells from p53-deficient mice. Oncogene 1993;8:3313-22. [PubMed]
- Serrano M, Lin AW, McCurrach ME, et al. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 1997;88:593-602. [Crossref] [PubMed]
- Dimri GP, Itahana K, Acosta M, et al. Regulation of a senescence checkpoint response by the E2F1 transcription factor and p14(ARF) tumor suppressor. Mol Cell Biol 2000;20:273-85. [Crossref] [PubMed]
- Damalas A, Kahan S, Shtutman M, et al. Deregulated beta-catenin induces a p53- and ARF-dependent growth arrest and cooperates with Ras in transformation. EMBO J 2001;20:4912-22. [Crossref] [PubMed]
- Post SM, Quintas-Cardama A, Terzian T, et al. p53-dependent senescence delays Emu-myc-induced B-cell lymphomagenesis. Oncogene 2010;29:1260-9. [Crossref] [PubMed]
- Noda A, Ning Y, Venable SF, et al. Cloning of senescent cell-derived inhibitors of DNA synthesis using an expression screen. Exp Cell Res 1994;211:90-8. [Crossref] [PubMed]
- Deng C, Zhang P, Harper JW, et al. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell 1995;82:675-84. [Crossref] [PubMed]
- Brugarolas J, Chandrasekaran C, Gordon JI, et al. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature 1995;377:552-7. [Crossref] [PubMed]
- Polyak K, Waldman T, He TC, et al. Genetic determinants of p53-induced apoptosis and growth arrest. Genes Dev 1996;10:1945-52. [Crossref] [PubMed]
- Fang L, Igarashi M, Leung J, et al. p21Waf1/Cip1/Sdi1 induces permanent growth arrest with markers of replicative senescence in human tumor cells lacking functional p53. Oncogene 1999;18:2789-97. [Crossref] [PubMed]
- Wang Y, Blandino G, Givol D. Induced p21waf expression in H1299 cell line promotes cell senescence and protects against cytotoxic effect of radiation and doxorubicin. Oncogene 1999;18:2643-9. [Crossref] [PubMed]
- Lowe SW, Ruley HE. Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev 1993;7:535-45. [Crossref] [PubMed]
- Debbas M, White E. Wild-type p53 mediates apoptosis by E1A, which is inhibited by E1B. Genes Dev 1993;7:546-54. [Crossref] [PubMed]
- Schmitt CA, McCurrach ME, de Stanchina E, et al. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev 1999;13:2670-7. [Crossref] [PubMed]
- Pierce AM, Gimenez-Conti IB, Schneider-Broussard R, et al. Increased E2F1 activity induces skin tumors in mice heterozygous and nullizygous for p53. Proc Natl Acad Sci 1998;95:8858-63. [Crossref] [PubMed]
- Symonds H, Krall L, Remington L, et al. p53-Dependent apoptosis suppresses tumor growth and progression in vivo. Cell 1994;78:703-11. [Crossref] [PubMed]
- Eischen CM, Roussel MF, Korsmeyer SJ, et al. Bax loss impairs Myc-induced apoptosis and circumvents the selection of p53 mutations during Myc-mediated lymphomagenesis. Mol Cell Biol 2001;21:7653-62. [Crossref] [PubMed]
- Hemann MT, Zilfou JT, Zhao Z, et al. Suppression of tumorigenesis by the p53 target PUMA. Proc Natl Acad Sci 2004;101:9333-8. [Crossref] [PubMed]
- Garrison SP, Jeffers JR, Yang C, et al. Selection against PUMA gene expression in Myc-driven B-cell lymphomagenesis. Mol Cell Biol 2008;28:5391-402. [Crossref] [PubMed]
- Michalak EM, Jansen E, Happo L, et al. Puma and to a lesser extent Noxa are suppressors of Myc-induced lymphomagenesis. Cell Death Differ 2009;16:684-96. [Crossref] [PubMed]
- Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell 2006;127:1323-34. [Crossref] [PubMed]
- Xue W, Zender L, Miething C, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 2007;445:656-60. [Crossref] [PubMed]
- Ventura A, Kirsch DG, McLaughlin ME, et al. Restoration of p53 function leads to tumour regression in vivo. Nature 2007;445:661-5. [Crossref] [PubMed]
- Li T, Kon N, Jiang L, et al. Tumor suppression in the absence of p53-mediated cell cycle arrest, apoptosis, and senescence. Cell 2012;149:1269-83. [Crossref] [PubMed]
- Valente LJ, Gray DH, Michalak EM, et al. p53 efficiently suppresses tumor development in the complete absence of its cell-cycle inhibitory and proapoptotic effectors p21, Puma, and Noxa. Cell Rep 2013;3:1339-45. [Crossref] [PubMed]
- Dixon SJ, Lemberg KM, Lamprecht MR, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 2012;149:1060-72. [Crossref] [PubMed]
- Jiang L, Kon N, Li T, et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature 2015;520:57-62. [Crossref] [PubMed]
- Maltzman W, Czyzyk L. UV irradiation stimulates levels of p53 cellular tumor antigen in nontransformed mouse cells. Mol Cell Biol 1984;4:1689-94. [Crossref] [PubMed]
- Kastan MB, Onyekwere O, Sidransky D, et al. Participation of p53 protein in the cellular response to DNA damage. Cancer Res 1991;51:6304-11. [PubMed]
- Fritsche M, Haessler C, Brandner G. Induction of nuclear accumulation of the tumor-suppressor protein p53 by DNA-damaging agents. Oncogene 1993;8:307-18. [PubMed]
- Zhan Q, Carrier F, Fornace AJ. Induction of cellular p53 activity by DNA-damaging agents and growth arrest. Mol Cell Biol 1993;13:4242-50. [Crossref] [PubMed]
- Lowe SW, Ruley HE, Jacks T, et al. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell 1993;74:957-67. [Crossref] [PubMed]
- Lowe SW, Jacks T, Housman DE, et al. Abrogation of oncogene-associated apoptosis allows transformation of p53-deficient cells. Proc Natl Acad Sci 1994;91:2026-30. [Crossref] [PubMed]
- Lowe SW, Schmitt EM, Smith SW, et al. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature 1993;362:847-9. [Crossref] [PubMed]
- Clarke AR, Purdie CA, Harrison DJ, et al. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature 1993;362:849-52. [Crossref] [PubMed]
- Lotem J, Sachs L. Hematopoietic cells from mice deficient in wild-type p53 are more resistant to induction of apoptosis by some agents. Blood 1993;82:1092-6. [PubMed]
- Merritt AJ, Potten CS, Kemp CJ, et al. The role of p53 in spontaneous and radiation-induced apoptosis in the gastrointestinal tract of normal and p53-deficient mice. Cancer Res 1994;54:614-7. [PubMed]
- Komarova EA, Christov K, Faerman AI, et al. Different impact of p53 and p21 on the radiation response of mouse tissues. Oncogene 2000;19:3791-8. [Crossref] [PubMed]
- Kastan MB, Zhan Q, el-Deiry WS, et al. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell 1992;71:587-97. [Crossref] [PubMed]
- Barlow C, Hirotsune S, Paylor R, et al. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell 1996;86:159-71. [Crossref] [PubMed]
- Elson A, Wang Y, Daugherty CJ, et al. Pleiotropic defects in ataxia-telangiectasia protein-deficient mice. Proc Natl Acad Sci 1996;93:13084-9. [Crossref] [PubMed]
- Xu Y, Baltimore D. Dual roles of ATM in the cellular response to radiation and in cell growth control. Genes Dev 1996;10:2401-10. [Crossref] [PubMed]
- Herzog KH, Chong MJ, Kapsetaki M, et al. Requirement for Atm in ionizing radiation-induced cell death in the developing central nervous system. Science 1998;280:1089-91. [Crossref] [PubMed]
- Gurley KE, Kemp CJ. Ataxia-telangiectasia mutated is not required for p53 induction and apoptosis in irradiated epithelial tissues. Mol Cancer Res 2007;5:1312-8. [Crossref] [PubMed]
- Cahilly-Snyder L, Yang-Feng T, Francke U, et al. Molecular analysis and chromosomal mapping of amplified genes isolated from a transformed mouse 3T3 cell line. Somat Cell Mol Genet 1987;13:235-44. [Crossref] [PubMed]
- Fakharzadeh SS, Trusko SP, George DL. Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. EMBO J 1991;10:1565-9. [PubMed]
- Oliner JD, Kinzler KW, Meltzer PS, et al. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 1992;358:80-3. [Crossref] [PubMed]
- Leach FS, Tokino T, Meltzer P, et al. p53 Mutation and MDM2 amplification in human soft tissue sarcomas. Cancer Res 1993;53:2231-4. [PubMed]
- Cordon-Cardo C, Latres E, Drobnjak M, et al. Molecular abnormalities of mdm2 and p53 genes in adult soft tissue sarcomas. Cancer Res 1994;54:794-9. [PubMed]
- Sheikh MS, Shao ZM, Hussain A, et al. The p53-binding protein MDM2 gene is differentially expressed in human breast carcinoma. Cancer Res 1993;53:3226-8. [PubMed]
- Reifenberger G, Liu L, Ichimura K, et al. Amplification and overexpression of the MDM2 gene in a subset of human malignant gliomas without p53 mutations. Cancer Res 1993;53:2736-9. [PubMed]
- Bueso-Ramos CE, Yang Y, deLeon E, et al. The human MDM-2 oncogene is overexpressed in leukemias. Blood 1993;82:2617-23. [PubMed]
- Watanabe T, Hotta T, Ichikawa A, et al. The MDM2 oncogene overexpression in chronic lymphocytic leukemia and low-grade lymphoma of B-cell origin. Blood 1994;84:3158-65. [PubMed]
- Matsumura T, Yoshihama Y, Kimura T, et al. p53 and MDM2 expression in oral squamous cell carcinoma. Oncology 2009;53:308-12. [Crossref] [PubMed]
- Oliner JD, Pietenpol JA, Thiagalingam S, et al. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature 1993;362:857-60. [Crossref] [PubMed]
- Chen J, Marechal V, Levine AJ. Mapping of the p53 and mdm-2 interaction domains. Mol Cell Biol 1993;13:4107-14. [Crossref] [PubMed]
- Finlay CA. The mdm-2 oncogene can overcome wild-type p53 suppression of transformed cell growth. Mol Cell Biol 1993;13:301-6. [Crossref] [PubMed]
- Chen CY, Oliner JD, Zhan Q, et al. Interactions between p53 and MDM2 in a mammalian cell cycle checkpoint pathway. Proc Natl Acad Sci 1994;91:2684-8. [Crossref] [PubMed]
- Haupt Y, Barak Y, Oren M. Cell type-specific inhibition of p53-mediated apoptosis by mdm2. EMBO J 1996;15:1596-606. [PubMed]
- Chen J, Wu X, Lin J, et al. mdm-2 inhibits the G1 arrest and apoptosis functions of the p53 tumor suppressor protein. Mol Cell Biol 1996;16:2445-52. [Crossref] [PubMed]
- Haupt Y, Maya R, Kazaz A, et al. Mdm2 promotes the rapid degradation of p53. Nature 1997;387:296-9. [Crossref] [PubMed]
- Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature 1997;387:299-303. [Crossref] [PubMed]
- Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett 1997;420:25-7. [Crossref] [PubMed]
- Fang S, Jensen JP, Ludwig RL, et al. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem 2000;275:8945-51. [Crossref] [PubMed]
- Boyd SD, Tsai KY, Jacks T. An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat Cell Biol 2000;2:563-8. [Crossref] [PubMed]
- Geyer RK, Yu ZK, Maki CG. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat Cell Biol 2000;2:569-73. [Crossref] [PubMed]
- Lohrum MA, Woods DB, Ludwig RL, et al. C-terminal ubiquitination of p53 contributes to nuclear export. Mol Cell Biol 2001;21:8521-32. [Crossref] [PubMed]
- Gu J, Nie L, Wiederschain D, et al. Identification of p53 sequence elements that are required for MDM2-mediated nuclear export. Mol Cell Biol 2001;21:8533-46. [Crossref] [PubMed]
- Li M, Brooks CL, Wu-Baer F, et al. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science 2003;302:1972-5. [Crossref] [PubMed]
- Rodriguez MS, Desterro JM, Lain S, et al. Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol Cell Biol 2000;20:8458-67. [Crossref] [PubMed]
- Nakamura S, Roth JA, Mukhopadhyay T. Multiple lysine mutations in the C-terminal domain of p53 interfere with MDM2-dependent protein degradation and ubiquitination. Mol Cell Biol 2000;20:9391-8. [Crossref] [PubMed]
- Ito A, Lai CH, Zhao X, et al. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J 2001;20:1331-40. [Crossref] [PubMed]
- Li M, Luo J, Brooks CL, et al. Acetylation of p53 inhibits its ubiquitination by Mdm2. J Biol Chem 2002;277:50607-11. [Crossref] [PubMed]
- Wu X, Bayle JH, Olson D, et al. The p53-mdm-2 autoregulatory feedback loop. Genes Dev 1993;7:1126-32. [Crossref] [PubMed]
- Juven T, Barak Y, Zauberman A, et al. Wild type p53 can mediate sequence-specific transactivation of an internal promoter within the mdm2 gene. Oncogene 1993;8:3411-6. [PubMed]
- Honda R, Yasuda H. Activity of MDM2, a ubiquitin ligase, toward p53 or itself is dependent on the RING finger domain of the ligase. Oncogene 2000;19:1473-6. [Crossref] [PubMed]
- Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 1995;378:203-6. [Crossref] [PubMed]
- Jones SN, Roe AE, Donehower LA, et al. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 1995;378:206-8. [Crossref] [PubMed]
- Jones SN, Sands AT, Hancock AR, et al. The tumorigenic potential and cell growth characteristics of p53-deficient cells are equivalent in the presence or absence of Mdm2. Proc Natl Acad Sci 1996;93:14106-11. [Crossref] [PubMed]
- Alt JR, Greiner TC, Cleveland JL, et al. Mdm2 haplo-insufficiency profoundly inhibits Myc-induced lymphomagenesis. EMBO J 2003;22:1442-50. [Crossref] [PubMed]
- Mendrysa SM, McElwee MK, Michalowski J, et al. mdm2 Is critical for inhibition of p53 during lymphopoiesis and the response to ionizing irradiation. Mol Cell Biol 2003;23:462-72. [Crossref] [PubMed]
- Jones SN, Hancock AR, Vogel H, et al. Overexpression of Mdm2 in mice reveals a p53-independent role for Mdm2 in tumorigenesis. Proc Natl Acad Sci 1998;95:15608-12. [Crossref] [PubMed]
- Wang P, Lushnikova T, Odvody J, et al. Elevated Mdm2 expression induces chromosomal instability and confers a survival and growth advantage to B cells. Oncogene 2008;27:1590-8. [Crossref] [PubMed]
- Shvarts A, Steegenga WT, Riteco N, et al. MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J 1996;15:5349-57. [PubMed]
- Shvarts A, Bazuine M, Dekker P, et al. Isolation and identification of the human homolog of a new p53-binding protein, Mdmx. Genomics 1997;43:34-42. [Crossref] [PubMed]
- Riemenschneider MJ, Knobbe CB, Reifenberger G. Refined mapping of 1q32 amplicons in malignant gliomas confirms MDM4 as the main amplification target. Int J Cancer 2003;104:752-7. [Crossref] [PubMed]
- Danovi D, Meulmeester E, Pasini D, et al. Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol Cell Biol 2004;24:5835-43. [Crossref] [PubMed]
- Bartel F, Schulz J, Bohnke A, et al. Significance of HDMX-S (or MDM4) mRNA splice variant overexpression and HDMX gene amplification on primary soft tissue sarcoma prognosis. Int J Cancer 2005;117:469-75. [Crossref] [PubMed]
- Laurie NA, Donovan SL, Shih CS, et al. Inactivation of the p53 pathway in retinoblastoma. Nature 2006;444:61-6. [Crossref] [PubMed]
- Gembarska A, Luciani F, Fedele C, et al. MDM4 is a key therapeutic target in cutaneous melanoma. Nat Med 2012;18:1239-47. [Crossref] [PubMed]
- Jackson MW, Berberich SJ. MdmX protects p53 from Mdm2-mediated degradation. Mol Cell Biol 2000;20:1001-7. [Crossref] [PubMed]
- Stad R, Ramos YF, Little N, et al. Hdmx stabilizes Mdm2 and p53. J Biol Chem 2000;275:28039-44. [PubMed]
- Parant J, Chavez-Reyes A, Little NA, et al. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet 2001;29:92-5. [Crossref] [PubMed]
- Migliorini D, Lazzerini Denchi E, Danovi D, et al. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol Cell Biol 2002;22:5527-38. [Crossref] [PubMed]
- Finch RA, Donoviel DB, Potter D, et al. mdmx is a negative regulator of p53 activity in vivo. Cancer Res 2002;62:3221-5. [PubMed]
- de Rozieres S, Maya R, Oren M, et al. The loss of mdm2 induces p53-mediated apoptosis. Oncogene 2000;19:1691-7. [Crossref] [PubMed]
- Steinman HA, Sluss HK, Sands AT, et al. Absence of p21 partially rescues Mdm4 loss and uncovers an antiproliferative effect of Mdm4 on cell growth. Oncogene 2004;23:303-6. [Crossref] [PubMed]
- Terzian T, Wang Y, Van Pelt CS, et al. Haploinsufficiency of Mdm2 and Mdm4 in tumorigenesis and development. Mol Cell Biol 2007;27:5479-85. [Crossref] [PubMed]
- Tanimura S, Ohtsuka S, Mitsui K, et al. MDM2 interacts with MDMX through their RING finger domains. FEBS Lett 1999;447:5-9. [Crossref] [PubMed]
- Sharp DA, Kratowicz SA, Sank MJ, et al. Stabilization of the MDM2 oncoprotein by interaction with the structurally related MDMX protein. J Biol Chem 1999;274:38189-96. [Crossref] [PubMed]
- Gu J, Kawai H, Nie L, et al. Mutual dependence of MDM2 and MDMX in their functional inactivation of p53. J Biol Chem 2002;277:19251-4. [Crossref] [PubMed]
- Linares LK, Hengstermann A, Ciechanover A, et al. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc Natl Acad Sci 2003;100:12009-14. [Crossref] [PubMed]
- Kawai H, Lopez-Pajares V, Kim MM, et al. RING domain-mediated interaction is a requirement for MDM2’s E3 ligase activity. Cancer Res 2007;67:6026-30. [Crossref] [PubMed]
- Wang X, Wang J, Jiang X. MdmX protein is essential for Mdm2 protein-mediated p53 polyubiquitination. J Biol Chem 2011;286:23725-34. [Crossref] [PubMed]
- Pan Y, Chen J. MDM2 promotes ubiquitination and degradation of MDMX. Mol Cell Biol 2003;23:5113-21. [Crossref] [PubMed]
- de Graaf P, Little NA, Ramos YF, et al. Hdmx protein stability is regulated by the ubiquitin ligase activity of Mdm2. J Biol Chem 2003;278:38315-24. [Crossref] [PubMed]
- Kawai H, Wiederschain D, Kitao H, et al. DNA damage-induced MDMX degradation is mediated by MDM2. J Biol Chem 2003;278:45946-53. [Crossref] [PubMed]
- Itahana K, Mao H, Jin A, et al. Targeted inactivation of Mdm2 RING finger E3 ubiquitin ligase activity in the mouse reveals mechanistic insights into p53 regulation. Cancer Cell 2007;12:355-66. [Crossref] [PubMed]
- Pant V, Xiong S, Iwakuma T, et al. Heterodimerization of Mdm2 and Mdm4 is critical for regulating p53 activity during embryogenesis but dispensable for p53 and Mdm2 stability. Proc Natl Acad Sci 2011;108:11995-2000. [Crossref] [PubMed]
- Huang L, Yan Z, Liao X, et al. The p53 inhibitors MDM2/MDMX complex is required for control of p53 activity in vivo. Proc Natl Acad Sci 2011;108:12001-6. [Crossref] [PubMed]
- Tollini LA, Jin A, Park J, et al. Regulation of p53 by Mdm2 E3 ligase function is dispensable in embryogenesis and development, but essential in response to DNA damage. Cancer Cell 2014;26:235-47. [Crossref] [PubMed]
- Zindy F, Eischen CM, Randle DH, et al. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev 1998;12:2424-33. [Crossref] [PubMed]
- de Stanchina E, McCurrach ME, Zindy F, et al. E1A signaling to p53 involves the p19(ARF) tumor suppressor. Genes Dev 1998;12:2434-42. [Crossref] [PubMed]
- Palmero I, Pantoja C, Serrano M. p19ARF links the tumour suppressor p53 to Ras. Nature 1998;395:125-6. [Crossref] [PubMed]
- Pomerantz J, Schreiber-Agus N, Liegeois NJ, et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell 1998;92:713-23. [Crossref] [PubMed]
- Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 1998;92:725-34. [Crossref] [PubMed]
- Kamijo T, Weber JD, Zambetti G, et al. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci 1998;95:8292-7. [Crossref] [PubMed]
- Stott FJ, Bates S, James MC, et al. The alternative product from the human CDKN2A locus, p14ARF, participates in a regulatory feedback loop with p53 and MDM2. EMBO J 1998;17:5001-14. [Crossref] [PubMed]
- Honda R, Yasuda H. Association of p19(ARF) with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J 1999;18:22-7. [Crossref] [PubMed]
- Midgley CA, Desterro JM, Saville MK, et al. An N-terminal p14ARF peptide blocks Mdm2-dependent ubiquitination in vitro and can activate p53 in vivo. Oncogene 2000;19:2312-23. [Crossref] [PubMed]
- Weber JD, Taylor LJ, Roussel MF, et al. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol 1999;1:20-6. [Crossref] [PubMed]
- Tao W, Levine AJ. P19ARF stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc Natl Acad Sci 1999;96:6937-41. [Crossref] [PubMed]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature 2009;461:1071-8. [Crossref] [PubMed]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell 2010;40:179-204. [Crossref] [PubMed]
- Meek DW, Anderson CW. Posttranslational modification of p53: cooperative integrators of function. Cold Spring Harb Perspect Biol 2009;1:a000950 [Crossref] [PubMed]
- Meek DW. Regulation of the p53 response and its relationship to cancer. Biochem J 2015;469:325-46. [Crossref] [PubMed]
- Lees-Miller SP, Sakaguchi K, Ullrich SJ, et al. Human DNA-activated protein kinase phosphorylates serines 15 and 37 in the amino-terminal transactivation domain of human p53. Mol Cell Biol 1992;12:5041-9. [Crossref] [PubMed]
- Banin S, Moyal L, Shieh S, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 1998;281:1674-7. [Crossref] [PubMed]
- Canman CE, Lim DS, Cimprich KA, et al. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 1998;281:1677-9. [Crossref] [PubMed]
- Tibbetts RS, Brumbaugh KM, Williams JM, et al. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev 1999;13:152-7. [Crossref] [PubMed]
- Shieh SY, Ahn J, Tamai K, et al. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev 2000;14:289-300. [PubMed]
- Hirao A, Kong YY, Matsuoka S, et al. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science 2000;287:1824-7. [Crossref] [PubMed]
- Saito S, Goodarzi AA, Higashimoto Y, et al. ATM mediates phosphorylation at multiple p53 sites, including Ser(46), in response to ionizing radiation. J Biol Chem 2002;277:12491-4. [Crossref] [PubMed]
- Dumaz N, Milne DM, Meek DW. Protein kinase CK1 is a p53-threonine 18 kinase which requires prior phosphorylation of serine 15. FEBS Lett 1999;463:312-6. [Crossref] [PubMed]
- Higashimoto Y, Saito S, Tong XH, et al. Human p53 is phosphorylated on serines 6 and 9 in response to DNA damage-inducing agents. J Biol Chem 2000;275:23199-203. [Crossref] [PubMed]
- Shieh SY, Ikeda M, Taya Y, et al. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 1997;91:325-34. [Crossref] [PubMed]
- Siliciano JD, Canman CE, Taya Y, et al. DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev 1997;11:3471-81. [Crossref] [PubMed]
- Unger T, Juven-Gershon T, Moallem E, et al. Critical role for Ser20 of human p53 in the negative regulation of p53 by Mdm2. EMBO J 1999;18:1805-14. [Crossref] [PubMed]
- Chehab NH, Malikzay A, Stavridi ES, et al. Phosphorylation of Ser-20 mediates stabilization of human p53 in response to DNA damage. Proc Natl Acad Sci 1999;96:13777-82. [Crossref] [PubMed]
- Sakaguchi K, Saito S, Higashimoto Y, et al. Damage-mediated phosphorylation of human p53 threonine 18 through a cascade mediated by a casein 1-like kinase. Effect on Mdm2 binding. J Biol Chem 2000;275:9278-83. [Crossref] [PubMed]
- Kussie PH, Gorina S, Marechal V, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 1996;274:948-53. [Crossref] [PubMed]
- Chao C, Hergenhahn M, Kaeser MD, et al. Cell type- and promoter-specific roles of Ser18 phosphorylation in regulating p53 responses. J Biol Chem 2003;278:41028-33. [Crossref] [PubMed]
- Sluss HK, Armata H, Gallant J, et al. Phosphorylation of serine 18 regulates distinct p53 functions in mice. Mol Cell Biol 2004;24:976-84. [Crossref] [PubMed]
- Loughery J, Cox M, Smith LM, et al. Critical role for p53-serine 15 phosphorylation in stimulating transactivation at p53-responsive promoters. Nucleic Acids Res 2014;42:7666-80. [Crossref] [PubMed]
- Sluss HK, Gannon H, Coles AH, et al. Phosphorylation of p53 serine 18 upregulates apoptosis to suppress Myc-induced tumorigenesis. Mol Cancer Res 2010;8:216-22. [Crossref] [PubMed]
- Wu Z, Earle J, Saito S, et al. Mutation of mouse p53 Ser23 and the response to DNA damage. Mol Cell Biol 2002;22:2441-9. [Crossref] [PubMed]
- MacPherson D, Kim J, Kim T, et al. Defective apoptosis and B-cell lymphomas in mice with p53 point mutation at Ser 23. EMBO J 2004;23:3689-99. [Crossref] [PubMed]
- Chao C, Herr D, Chun J, et al. Ser18 and 23 phosphorylation is required for p53-dependent apoptosis and tumor suppression. EMBO J 2006;25:2615-22. [PubMed]
- Chen L, Gilkes DM, Pan Y, et al. ATM and Chk2-dependent phosphorylation of MDMX contribute to p53 activation after DNA damage. EMBO J 2005;24:3411-22. [Crossref] [PubMed]
- Okamoto K, Kashima K, Pereg Y, et al. DNA damage-induced phosphorylation of MdmX at serine 367 activates p53 by targeting MdmX for Mdm2-dependent degradation. Mol Cell Biol 2005;25:9608-20. [Crossref] [PubMed]
- LeBron C, Chen L, Gilkes DM, et al. Regulation of MDMX nuclear import and degradation by Chk2 and 14-3-3. EMBO J 2006;25:1196-206. [Crossref] [PubMed]
- Pereg Y, Lam S, Teunisse A, et al. Differential roles of ATM- and Chk2-mediated phosphorylations of Hdmx in response to DNA damage. Mol Cell Biol 2006;26:6819-31. [Crossref] [PubMed]
- Pereg Y, Shkedy D, de Graaf P, et al. Phosphorylation of Hdmx mediates its Hdm2- and ATM-dependent degradation in response to DNA damage. Proc Natl Acad Sci 2005;102:5056-61. [Crossref] [PubMed]
- Meulmeester E, Maurice MM, Boutell C, et al. Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol Cell 2005;18:565-76. [Crossref] [PubMed]
- Wang YV, Leblanc M, Wade M, et al. Increased radio-resistance and accelerated B-cell lymphomas in mice with Mdmx mutations that prevent modifications by DNA damage-activated kinases. Cancer Cell 2009;16:33-43. [Crossref] [PubMed]
- Khosravi R, Maya R, Gottlieb T, et al. Rapid ATM-dependent phosphorylation of MDM2 precedes p53 accumulation in response to DNA damage. Proc Natl Acad Sci 1999;96:14973-7. [Crossref] [PubMed]
- Maya R, Balass M, Kim ST, et al. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev 2001;15:1067-77. [Crossref] [PubMed]
- Lu X, Ma O, Nguyen TA, et al. The Wip1 Phosphatase acts as a gatekeeper in the p53-Mdm2 autoregulatory loop. Cancer Cell 2007;12:342-54. [Crossref] [PubMed]
- Stommel JM, Wahl GM. Accelerated MDM2 auto-degradation induced by DNA-damage kinases is required for p53 activation. EMBO J 2004;23:1547-56. [Crossref] [PubMed]
- Cheng Q, Chen L, Li Z, et al. ATM activates p53 by regulating MDM2 oligomerization and E3 processivity. EMBO J 2009;28:3857-67. [Crossref] [PubMed]
- Cheng Q, Cross B, Li B, et al. Regulation of MDM2 E3 ligase activity by phosphorylation after DNA damage. Mol Cell Biol 2011;31:4951-63. [Crossref] [PubMed]
- Gannon HS, Woda BA, Jones SN. ATM phosphorylation of Mdm2 Ser394 regulates the amplitude and duration of the DNA damage response in mice. Cancer Cell 2012;21:668-79. [Crossref] [PubMed]
- Carr MI, Roderick JE, Gannon HS, et al. Mdm2 Phosphorylation Regulates Its Stability and Has Contrasting Effects on Oncogene and Radiation-Induced Tumorigenesis. Cell Rep 2016;16:2618-29. [Crossref] [PubMed]
- Kharbanda S, Ren R, Pandey P, et al. Activation of the c-Abl tyrosine kinase in the stress response to DNA-damaging agents. Nature 1995;376:785-8. [Crossref] [PubMed]
- Liu ZG, Baskaran R, Lea-Chou ET, et al. Three distinct signalling responses by murine fibroblasts to genotoxic stress. Nature 1996;384:273-6. [Crossref] [PubMed]
- Wang X, Zeng L, Wang J, et al. A positive role for c-Abl in Atm and Atr activation in DNA damage response. Cell Death Differ 2011;18:5-15. [Crossref] [PubMed]
- Baskaran R, Wood LD, Whitaker LL, et al. Ataxia telangiectasia mutant protein activates c-Abl tyrosine kinase in response to ionizing radiation. Nature 1997;387:516-9. [Crossref] [PubMed]
- Shafman T, Khanna KK, Kedar P, et al. Interaction between ATM protein and c-Abl in response to DNA damage. Nature 1997;387:520-3. [Crossref] [PubMed]
- Sawyers CL, McLaughlin J, Goga A, et al. The nuclear tyrosine kinase c-Abl negatively regulates cell growth. Cell 1994;77:121-31. [Crossref] [PubMed]
- Wen ST, Jackson PK, Van Etten RA. The cytostatic function of c-Abl is controlled by multiple nuclear localization signals and requires the p53 and Rb tumor suppressor gene products. EMBO J 1996;15:1583-95. [PubMed]
- Yuan ZM, Huang Y, Whang Y, et al. Role for c-Abl tyrosine kinase in growth arrest response to DNA damage. Nature 1996;382:272-4. [Crossref] [PubMed]
- Yuan ZM, Huang Y, Ishiko T, et al. Regulation of DNA damage-induced apoptosis by the c-Abl tyrosine kinase. Proc Natl Acad Sci 1997;94:1437-40. [Crossref] [PubMed]
- Gong JG, Costanzo A, Yang HQ, et al. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature 1999;399:806-9. [Crossref] [PubMed]
- Agami R, Blandino G, Oren M, et al. Interaction of c-Abl and p73alpha and their collaboration to induce apoptosis. Nature 1999;399:809-13. [Crossref] [PubMed]
- Yuan ZM, Shioya H, Ishiko T, et al. p73 is regulated by tyrosine kinase c-Abl in the apoptotic response to DNA damage. Nature 1999;399:814-7. [Crossref] [PubMed]
- Zuckerman V, Lenos K, Popowicz GM, et al. c-Abl phosphorylates Hdmx and regulates its interaction with p53. J Biol Chem 2009;284:4031-9. [Crossref] [PubMed]
- Sionov RV, Moallem E, Berger M, et al. c-Abl neutralizes the inhibitory effect of Mdm2 on p53. J Biol Chem 1999;274:8371-4. [Crossref] [PubMed]
- Sionov RV, Coen S, Goldberg Z, et al. c-Abl regulates p53 levels under normal and stress conditions by preventing its nuclear export and ubiquitination. Mol Cell Biol 2001;21:5869-78. [Crossref] [PubMed]
- Goldberg Z, Vogt Sionov R, Berger M, et al. Tyrosine phosphorylation of Mdm2 by c-Abl: implications for p53 regulation. EMBO J 2002;21:3715-27. [Crossref] [PubMed]
- Dias SS, Milne DM, Meek DW. c-Abl phosphorylates Hdm2 at tyrosine 276 in response to DNA damage and regulates interaction with ARF. Oncogene 2006;25:6666-71. [Crossref] [PubMed]
- Waning DL, Lehman JA, Batuello CN, et al. c-Abl phosphorylation of Mdm2 facilitates Mdm2-Mdmx complex formation. J Biol Chem 2011;286:216-22. [Crossref] [PubMed]
- Hay TJ, Meek DW. Multiple sites of in vivo phosphorylation in the MDM2 oncoprotein cluster within two important functional domains. FEBS Lett 2000;478:183-6. [Crossref] [PubMed]
- Hjerrild M, Milne D, Dumaz N, et al. Phosphorylation of murine double minute clone 2 (MDM2) protein at serine-267 by protein kinase CK2 in vitro and in cultured cells. Biochem J 2001;355:347-56. [Crossref] [PubMed]
- Winter M, Milne D, Dias S, et al. Protein kinase CK1delta phosphorylates key sites in the acidic domain of murine double-minute clone 2 protein (MDM2) that regulate p53 turnover. Biochemistry 2004;43:16356-64. [Crossref] [PubMed]
- Allende-Vega N, Dias S, Milne D, et al. Phosphorylation of the acidic domain of Mdm2 by protein kinase CK2. Mol Cell Biochem 2005;274:85-90. [Crossref] [PubMed]
- Kulikov R, Boehme KA, Blattner C. Glycogen synthase kinase 3-dependent phosphorylation of Mdm2 regulates p53 abundance. Mol Cell Biol 2005;25:7170-80. [Crossref] [PubMed]
- Blattner C, Hay T, Meek DW, et al. Hypophosphorylation of Mdm2 augments p53 stability. Mol Cell Biol 2002;22:6170-82. [Crossref] [PubMed]
- Boehme KA, Kulikov R, Blattner C. p53 stabilization in response to DNA damage requires Akt/PKB and DNA-PK. Proc Natl Acad Sci 2008;105:7785-90. [Crossref] [PubMed]
- Mayo LD, Donner DB. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc Natl Acad Sci 2001;98:11598-603. [Crossref] [PubMed]
- Zhou BP, Liao Y, Xia W, et al. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat Cell Biol 2001;3:973-82. [Crossref] [PubMed]
- Feng J, Tamaskovic R, Yang Z, et al. Stabilization of Mdm2 via decreased ubiquitination is mediated by protein kinase B/Akt-dependent phosphorylation. J Biol Chem 2004;279:35510-7. [Crossref] [PubMed]
- Momand J, Zambetti GP, Olson DC, et al. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 1992;69:1237-45. [Crossref] [PubMed]
- Linares LK, Kiernan R, Triboulet R, et al. Intrinsic ubiquitination activity of PCAF controls the stability of the oncoprotein Hdm2. Nat Cell Biol 2007;9:331-8. [Crossref] [PubMed]
- He Y, Tollini L, Kim TH, et al. The anaphase-promoting complex/cyclosome is an E3 ubiquitin ligase for Mdm2. Cell Cycle 2014;13:2101-9. [Crossref] [PubMed]
- Inuzuka H, Tseng A, Gao D, et al. Phosphorylation by casein kinase I promotes the turnover of the Mdm2 oncoprotein via the SCF(beta-TRCP) ubiquitin ligase. Cancer Cell 2010;18:147-59. [Crossref] [PubMed]
- Malonia SK, Dutta P, Santra MK, et al. F-box protein FBXO31 directs degradation of MDM2 to facilitate p53-mediated growth arrest following genotoxic stress. Proc Natl Acad Sci 2015;112:8632-7. [Crossref] [PubMed]
- Wang Z, Inuzuka H, Zhong J, et al. DNA damage-induced activation of ATM promotes beta-TRCP-mediated Mdm2 ubiquitination and destruction. Oncotarget 2012;3:1026-35. [Crossref] [PubMed]
- Buschmann T, Lerner D, Lee CG, et al. The Mdm-2 amino terminus is required for Mdm2 binding and SUMO-1 conjugation by the E2 SUMO-1 conjugating enzyme Ubc9. J Biol Chem 2001;276:40389-95. [Crossref] [PubMed]
- Watson IR, Li BK, Roche O, et al. Chemotherapy induces NEDP1-mediated destabilization of MDM2. Oncogene 2010;29:297-304. [Crossref] [PubMed]
- Li M, Chen D, Shiloh A, et al. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 2002;416:648-53. [Crossref] [PubMed]
- Li M, Brooks CL, Kon N, et al. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell 2004;13:879-86. [Crossref] [PubMed]
- Tang J, Qu LK, Zhang J, et al. Critical role for Daxx in regulating Mdm2. Nat Cell Biol 2006;8:855-62. [Crossref] [PubMed]
- Tang J, Agrawal T, Cheng Q, et al. Phosphorylation of Daxx by ATM contributes to DNA damage-induced p53 activation. PLoS One 2013;8:e55813 [Crossref] [PubMed]
- Stevenson LF, Sparks A, Allende-Vega N, et al. The deubiquitinating enzyme USP2a regulates the p53 pathway by targeting Mdm2. EMBO J 2007;26:976-86. [Crossref] [PubMed]
- Zou Q, Jin J, Hu H, et al. USP15 stabilizes MDM2 to mediate cancer-cell survival and inhibit antitumor T cell responses. Nat Immunol 2014;15:562-70. [Crossref] [PubMed]
- Allende-Vega N, Sparks A, Lane DP, et al. MdmX is a substrate for the deubiquitinating enzyme USP2a. Oncogene 2010;29:432-41. [Crossref] [PubMed]
- Roth JA, Nguyen D, Lawrence DD, et al. Retrovirus-mediated wild-type p53 gene transfer to tumors of patients with lung cancer. Nat Med 1996;2:985-91. [Crossref] [PubMed]
- Jiang M, Milner J. Selective silencing of viral gene expression in HPV-positive human cervical carcinoma cells treated with siRNA, a primer of RNA interference. Oncogene 2002;21:6041-8. [Crossref] [PubMed]
- Yu Y, Sun P, Sun L, et al. Downregulation of MDM2 expression by RNAi inhibits LoVo human colorectal adenocarcinoma cells growth and the treatment of LoVo cells with mdm2siRNA3 enhances the sensitivity to cisplatin. Biochem Biophys Res Commun 2006;339:71-8. [Crossref] [PubMed]
- Mayordomo JI, Loftus DJ, Sakamoto H, et al. Therapy of murine tumors with p53 wild-type and mutant sequence peptide-based vaccines. J Exp Med 1996;183:1357-65. [Crossref] [PubMed]
- Vassilev LT, Vu BT, Graves B, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 2004;303:844-8. [Crossref] [PubMed]
- Grasberger BL, Lu T, Schubert C, et al. Discovery and cocrystal structure of benzodiazepinedione HDM2 antagonists that activate p53 in cells. J Med Chem 2005;48:909-12. [Crossref] [PubMed]
- Ding K, Lu Y, Nikolovska-Coleska Z, et al. Structure-based design of spiro-oxindoles as potent, specific small-molecule inhibitors of the MDM2-p53 interaction. J Med Chem 2006;49:3432-5. [Crossref] [PubMed]
- Karni-Schmidt O, Lokshin M, Prives C. The Roles of MDM2 and MDMX in Cancer. Annu Rev Pathol 2016;11:617-44. [Crossref] [PubMed]
- Hu B, Gilkes DM, Farooqi B, et al. MDMX overexpression prevents p53 activation by the MDM2 inhibitor Nutlin. J Biol Chem 2006;281:33030-5. [Crossref] [PubMed]
- Patton JT, Mayo LD, Singhi AD, et al. Levels of HdmX expression dictate the sensitivity of normal and transformed cells to Nutlin-3. Cancer Res 2006;66:3169-76. [Crossref] [PubMed]
- Popowicz GM, Czarna A, Wolf S, et al. Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery. Cell Cycle 2010;9:1104-11. [Crossref] [PubMed]
- Reed D, Shen Y, Shelat AA, et al. Identification and characterization of the first small molecule inhibitor of MDMX. J Biol Chem 2010;285:10786-96. [Crossref] [PubMed]
- Wang H, Ma X, Ren S, et al. A small-molecule inhibitor of MDMX activates p53 and induces apoptosis. Mol Cancer Ther 2011;10:69-79. [Crossref] [PubMed]
- Vaseva AV, Yallowitz AR, Marchenko ND, et al. Blockade of Hsp90 by 17AAG antagonizes MDMX and synergizes with Nutlin to induce p53-mediated apoptosis in solid tumors. Cell Death Dis 2011;2:e156 [Crossref] [PubMed]
- Graves B, Thompson T, Xia M, et al. Activation of the p53 pathway by small-molecule-induced MDM2 and MDMX dimerization. Proc Natl Acad Sci USA 2012;109:11788-93. [Crossref] [PubMed]
- Issaeva N, Bozko P, Enge M. Pet al. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med 2004;10:1321-8. [Crossref] [PubMed]
- Nieves-Neira W, Rivera MI, Kohlhagen G, et al. DNA protein cross-links produced by NSC 652287, a novel thiophene derivative active against human renal cancer cells. Mol Pharmacol 1999;56:478-84. [PubMed]
- Rivera MI, Stinson SF, Vistica DT, et al. Selective toxicity of the tricyclic thiophene NSC 652287 in renal carcinoma cell lines: differential accumulation and metabolism. Biochem Pharmacol 1999;57:1283-95. [Crossref] [PubMed]
- Di Conza G, Buttarelli M, Monti O, et al. IGF-1R/MDM2 relationship confers enhanced sensitivity to RITA in Ewing sarcoma cells. Mol Cancer Ther 2012;11:1247-56. [Crossref] [PubMed]
- Bernal F, Tyler AF, Korsmeyer SJ, et al. Reactivation of the p53 tumor suppressor pathway by a stapled p53 peptide. J Am Chem Soc 2007;129:2456-7. [Crossref] [PubMed]
- Bernal F, Wade M, Godes M, et al. A stapled p53 helix overcomes HDMX-mediated suppression of p53. Cancer Cell 2010;18:411-22. [Crossref] [PubMed]
- Schafmeister CE, Po J, Verdine GL. An All-Hydrocarbon Cross-Linking System for Enhancing the Helicity and Metabolic Stability of Peptides. J Am Chem Soc 2000;122:5891-2. [Crossref]
- Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer 2013;13:83-96. [Crossref] [PubMed]
- Yang Y, Ludwig RL, Jensen JP, et al. Small molecule inhibitors of HDM2 ubiquitin ligase activity stabilize and activate p53 in cells. Cancer Cell 2005;7:547-59. [Crossref] [PubMed]
- Roxburgh P, Hock AK, Dickens MP, et al. Small molecules that bind the Mdm2 RING stabilize and activate p53. Carcinogenesis 2012;33:791-8. [Crossref] [PubMed]
- Herman AG, Hayano M, Poyurovsky MV, et al. Discovery of Mdm2-MdmX E3 ligase inhibitors using a cell-based ubiquitination assay. Cancer Discov 2011;1:312-25. [Crossref] [PubMed]
- Carvajal D, Tovar C, Yang H, et al. Activation of p53 by MDM2 antagonists can protect proliferating cells from mitotic inhibitors. Cancer Res 2005;65:1918-24. [Crossref] [PubMed]
- Kranz D, Dobbelstein M. Nongenotoxic p53 activation protects cells against S-phase-specific chemotherapy. Cancer Res 2006;66:10274-80. [Crossref] [PubMed]
- Komarov PG, Komarova EA, Kondratov RV, et al. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science 1999;285:1733-7. [Crossref] [PubMed]
- Ganapathy S, Xiao S, Seo SJ, et al. Low-dose arsenic induces chemotherapy protection via p53/NF-kappaB-mediated metabolic regulation. Oncogene 2014;33:1359-66. [Crossref] [PubMed]
- Ganapathy S, Xiao S, Yang M, et al. A low-dose arsenic-induced p53 protein-mediated metabolic mechanism of radiotherapy protection. J Biol Chem 2014;289:5340-7. [Crossref] [PubMed]
- Su H, Ganapathy S, Li X, et al. p53-based strategy for protection of bone marrow from Y-90 ibritumomab tiuxetan. Int J Radiat Oncol Biol Phys 2015;92:1116-22. [Crossref] [PubMed]
- Ha CS, Michalek JE, Elledge R, et al. p53-based strategy to reduce hematological toxicity of chemotherapy: A proof of principle study. Mol Oncol 2016;10:148-56. [Crossref] [PubMed]


