
Mutant p53 talks to proteasomes—is there a feedback loop between Nrf2 and mutant p53?
The p53 protein is a powerful tumor suppressor, often termed as ‘guardian of the genome’, whose function is lost in many types of human cancers (1). The loss of function is impelled either by de-regulation of the E3 ubiquitin ligase, MDM2 protein (2) or by the TP53 gene mutations of missense type (3). Mutations in TP53 gene occur at a very high frequency and range from 10% to 97% depending on the cancer type (4). Single amino acid substitutions result in the loss of tumor suppression transcription function, promote the change of the conformation and loss of the sequence-specific DNA binding function of mutant p53 (mtp53) (5).
The mutations in the TP53 gene not only terminate the wild-type activity of p53, but also foster gaining of new functions (GOFs) by p53 mutants. In healthy tissue, p53 regulates several hundreds of genes that are pivotal in sufficient functioning of the DNA repair pathway, apoptosis, cell cycle arrest and maintaining cell homeostasis. GOFs by mtp53 evolve to enable sustained growth of cancer cells through enhancing nutrient uptake, synthesis of building blocks or fueling the pool of NAD(P)+ that are essential for cancer cells to divide, migrate and become resistant to treatment. Therefore, p53 mutations are invariably linked with worse prognosis and shorter overall survival of cancer patients.
Until recently, it was believed that mtp53 protein accumulates only in cancerous tissue (6). This was however put into question, as elevated levels of R172H mtp53 were found in proliferating cells of normal tissues of adult p53R172H/R172H knock-in mice, with no sign of tumor transformation and at the levels similar to wild-type p53 (7). It is known that wild-type p53 accumulates upon activation by ionizing radiation (8) but mtp53 accumulated more rapidly in the mouse knock-in model and remained at elevated levels longer than wild-type p53. This implies that similar mechanisms govern the turnover of wild-type and mutant p53 proteins, however, mtp53 accumulation is restricted to proliferating cells only. Albeit, around 2,000 mutations in the TP53 gene were described as well as the various mtp53-GOFs phenotypes, we are still on the way to divulge the mechanism underlying the emergence of certain mtp53 GOFs. Several studies ascribed these GOFs to the interactions of mtp53 with other transcription factors including NF-Y, SREBF2, ETS2 or ΔNp63. In addition, mtp53 binds to and inhibits other p53 family members like, TAp63 and TAp73 or wild-type p53 and interferes with miRNA biogenesis (9).
The structural and functional diversity between different p53 mutants brings about high variance in the mechanisms how p53 mutants utilize their pro-oncogenic signatures and in result, making it very challenging to apply mtp53 as a predictive factor for the treatment outcome in cancer therapies. Even though on-going efforts, it still remains of the outmost importance to determine the outcome of the mtp53 gain of new functions across different tumor types.
In August issue of Nature Cell Biology, Walerych et al. (10) addressed this challenge and singled out a universal mtp53 GOFs program that is an explicit transcriptional signature executing the expression of the proteasome machinery genes.
To seek for the major common target of cancer-related p53 variants, Walerych et al. (10,11) utilized a multi-omics approach converging on the whole-cell proteome and chip-sequencing analyses done in triple negative breast cancer (TNBC) cell line, MDA-MB-231 before and after silencing of mtp53. The analyses depicted a 72-gene integrated signature, which after pathway analysis, identified the proteasome-ubiquitination pathway to be the most changed upon mtp53 depletion. Next, comparison of MDA-MB-231 transcriptome with mtp53 mRNA profiles in four other TNBC cell lines harbouring cancer-related p53 mutants, revealed a signature of 205 common genes that in accordance with previous analysis, was most significantly enriched with genes coding for components of 26S proteasome. To validate these findings in more clinical setting, the authors performed a patient-based analysis in which they found that proteasome gene signature associated with poor prognosis and with the mtp53 status in breast cancer patients. Consequently, mtp53 was attributed with the enhanced proteasomal activity as measured in cancer cell lines of various origins and in studies in R172H knock-in mice.
The transcriptional regulation of 37 genes coding 26S proteasome and immunoproteasome subunits, found among the 72- and 205-gene group, was attributed to the interaction of p53 mutants with Nrf2 [NF-E2-related factor 2 (NFE2L2)], a pleiotropic transcription factor that regulates several different antioxidant pathways (12). Nrf2 is known to support the synthesis and regeneration of glutathione (GSH) and the production of NADPH. It is upregulated in cancers in consequence to loss of function mutations in Keap-1 and high levels of Nrf2 are associated with cisplatin resistance (13). Next, Nrf2 was shown to be upregulated by mtp53 and the mechanism is possibly via the cross-talk with Sp1 transcription that binds to NFE2L2 gene promoter (14).
The thorough analysis by Walerych and coworkers, demonstrates that Nrf2 engages mtp53s to the proteasomes’ genes, conversely to wtp53. Fluorescence microscopy analysis further corroborated the idea that mtp53s co-opt with Nrf2 to drive the expression of proteasome genes, as mtp53 co-localizes with Nrf2 in the nucleus and interacts with Nrf2 in the nuclear fraction. Next, the affinity of Nrf2 to GOFs mtp53 protein was supported in immunoprecipitation studies after treatment of cells with PRIMA-1MET/APR-246, a molecule that was shown to restore wild-type conformation to mtp53 and to induce p53-related apoptosis (15). Only mutant and not APR-246 activated wild-type p53 binds Nrf2. Hence, silencing of Nrf2 and mtp53 reduces the mRNA levels of the 26S proteasome genes. Interestingly, mtp53 has opposing effect on other Nrf2 target gene HMOX-1 encoding heme-oxygenase (HO-1) (16), an enzyme involved in maintaining iron pool and free radicals clearance. This implies at least dual function of Nrf2-mtp53 axis in cancer cells. One would be attributed to regulating the expression of proteasome genes and the second to regulating the classical, Nrf2 anti-oxidant response, though with the opposing outcome. Thus, although the up-regulation of proteasomal function has a clear survival advantage in cancer cells, downregulation of HO-1 still remains an unresolved puzzle. It is likely that other factors co-opt with Nrf2 and mtp53, to regulate the expression of HMXO-1 in mtp53 cancer cells or that the cellular effect that HO-1 exerts has already been taken over by other enzyme/pathway. Finally, it cannot be excluded that to study how Nrf2 regulates the expression of HMXO-1 in mtp53 cancer cells, more hostile conditions, mimicking tumor environment need to be applied.
Ubiquitin-proteasome system (UPS) is the major mechanism by which the proteins are degraded and as such is a pivotal player in protein homeostasis (proteostasis). High incidence of gene mutations and aneuploidy in cancers lead to accumulation of mutated proteins that often fail in proper folding. Multiple evidence implies that cancer cells are dependent on UPS for the timely removal of accumulated, unfolded or aggregated proteins in order to escape the so-called ‘proteotoxic crisis’ and this has already been utilized in cancer treatment. Thus, the discovery of the proteasome inhibitor bortezomib provided the first direct evidence that it is possible to inhibit the UPS and is now exploited in the therapy for multiple myeloma (17). The efficiency of proteasomal inhibitors in certain tumor types is however, questionable. Hence, as mtp53s co-opt with Nrf2 to up-regulate UPS, Walerych et al. further demonstrated, that mtp53-harbouring breast cancer cells are resistant to carfilzomib, novel proteasomal inhibitor and that the effect was dependent on mtp53. The resistance was due to the activation of the so-called ‘bounce-back’ effect where mtp53 up-regulates 26S proteasome genes transcription after treatment with carfilzomib (Figure 1). The finding that Nrf2 co-immunoprecipitated with mtp53s but not with wild-type p53 protein, prompted authors to investigate if inhibition of GOFs mtp53 could overcome the resistance to carfilzomib. Thus, they showed that APR-246, a compound currently in phase I/II clinical trials, abolishes the resistance of TNBC cells to carfilzomib in mouse xenograft model. This constitutes promising grounds for further investigation of APR-246 in combination with proteasomal inhibitor in a more clinical setting.
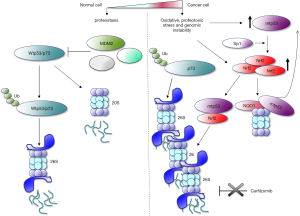
26S proteasome is composed of 20S catalytic subunit that at its end is capped with 19S regulatory particles (Figure 1). The degradation by 26S proteasome is mediated by the ubiquitin system and p53 is one of its best-studied substrate. However, ubiquitin-independent degradation by 20S proteasome has also been demonstrated to regulate p53 and other tumor suppressors turnover and was found de-regulated in cancers.
Nrf2 target, NAD(P)H:quinone oxidoreductase (NQO1), is an enzyme that reduces various quinones and is inhibited by the anticoagulant dicoumarol. NQO1 binds to p53 and TAp73 and stabilizes them by inhibiting the degradation by 20S proteasome (18) that is reversed by dicoumarol. TAp73 is also stabilized in cancer cells by a specific 20S proteasome inhibitor, withaferin A and the effect is due to the enhanced binding of TAp73 to NQO1 that only occurs upon the oxidative insult (19). Of note, it was demonstrated that R175H, R248H, and R273H mtp53s are resistant to dicoumarol-induced 20S degradation (20) and that the resistance is mediated by the enhanced binding of mtp53 to NQO1. This mechanism could at least partially contribute to the stabilization of mtp53 in cancer cells.
Hence, as also commented by Oren and Kotler (11), the story on how mtp53 co-opts with Nrf2 to talk to proteasome is likely to continue to complete the picture. To this end, there seems to be a feedback loop between the transcriptional upregulation of Nrf2 by mtp53 and Nrf2-NQO1-mediated stabilization of unfolded mtp53 protein. Yet, since different group of genes were found downregulated in TNBC upon different mtp53 silencing, it appears unlikely that the transcription-mediated regulation of proteasome machinery would be the only common mechanism underlying the GOFs phenotype. Studies divulging the interplay between ROS-scavenging enzymes and mtp53 GOFs phenotypes should advance understanding of GOFs phenotypes for better stratification of mtp53-harbouring patients to treatment.
Acknowledgments
Funding: This work was supported by KI/MD Anderson collaborative grant (Strategic Research Programme in Cancer, StratCan, KI) and Åke Wibergs Stiftelse (M14-0311).
Footnote
Provenance: This is a guest Commentary commissioned by guest editor Zhi-Min Yuan, MD, PhD (Department of Genetics and Complex Diseases, Harvard University School of Public Health, Boston, MA, USA).
Provenance and Peer Review: This article was commissioned by the Guest Editor (Zhi-Min Yuan) for the series “p53 Biology and Cancer” published in Translational Cancer Research. The article did not undergo external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.11.73). The series “p53 Biology and Cancer” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lane DP. Cancer. p53, guardian of the genome. Nature 1992;358:15-6. [Crossref] [PubMed]
- Haupt Y, Maya R, Kazaz A, et al. Mdm2 promotes the rapid degradation of p53. Nature 1997;387:296-9. [Crossref] [PubMed]
- Soussi T, Wiman KG. TP53: an oncogene in disguise. Cell Death Differ 2015;22:1239-49. [Crossref] [PubMed]
- Rivlin N, Brosh R, Oren M, et al. Mutations in the p53 Tumor Suppressor Gene: Important Milestones at the Various Steps of Tumorigenesis. Genes Cancer 2011;2:466-74. [Crossref] [PubMed]
- Dittmer D, Pati S, Zambetti G, et al. Gain of function mutations in p53. Nat Genet 1993;4:42-6. [Crossref] [PubMed]
- Terzian T, Suh YA, Iwakuma T, et al. The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss. Genes Dev 2008;22:1337-44. [Crossref] [PubMed]
- Goh AM, Xue Y, Leushacke M, et al. Mutant p53 accumulates in cycling and proliferating cells in the normal tissues of p53 R172H mutant mice. Oncotarget 2015;6:17968-80. [Crossref] [PubMed]
- Maya R, Balass M, Kim ST, et al. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev 2001;15:1067-77. [Crossref] [PubMed]
- Garibaldi F, Falcone E, Trisciuoglio D, et al. Mutant p53 inhibits miRNA biogenesis by interfering with the microprocessor complex. Oncogene 2016;35:3760-70. [Crossref] [PubMed]
- Walerych D, Lisek K, Sommaggio R, et al. Proteasome machinery is instrumental in a common gain-of-function program of the p53 missense mutants in cancer. Nat Cell Biol 2016;18:897-909. [Crossref] [PubMed]
- Oren M, Kotler E. p53 mutations promote proteasomal activity. Nat Cell Biol 2016;18:833-5. [Crossref] [PubMed]
- Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov 2013;12:931-47. [Crossref] [PubMed]
- Homma S, Ishii Y, Morishima Y, et al. Nrf2 enhances cell proliferation and resistance to anticancer drugs in human lung cancer. Clin Cancer Res 2009;15:3423-32. [Crossref] [PubMed]
- Tung MC, Lin PL, Wang YC, et al. Mutant p53 confers chemoresistance in non-small cell lung cancer by upregulating Nrf2. Oncotarget 2015;6:41692-705. [PubMed]
- Lambert JM, Gorzov P, Veprintsev DB. PRIMA-1 reactivates mutant p53 by covalent binding to the core domain. Cancer Cell 2009;15:376-88. [Crossref] [PubMed]
- Kalo E, Kogan-Sakin I, Solomon H, et al. Mutant p53R273H attenuates the expression of phase 2 detoxifying enzymes and promotes the survival of cells with high levels of reactive oxygen species. J Cell Sci 2012;125:5578-86. [Crossref] [PubMed]
- Richardson PG, Sonneveld P, Schuster MW, et al. Bortezomib or high-dose dexamethasone for relapsed multiple myeloma. N Engl J Med 2005;352:2487-98. [Crossref] [PubMed]
- Asher G, Tsvetkov P, Kahana C, et al. A mechanism of ubiquitin-independent proteasomal degradation of the tumor suppressors p53 and p73. Genes Dev 2005;19:316-21. [Crossref] [PubMed]
- Kostecka A, Sznarkowska A, Meller K, et al. JNK-NQO1 axis drives TAp73-mediated tumor suppression upon oxidative and proteasomal stress. Cell Death Dis 2014;5:e1484 [Crossref] [PubMed]
- Asher G, Lotem J, Tsvetkov P, et al. P53 hot-spot mutants are resistant to ubiquitin-independent degradation by increased binding to NAD(P)H:quinone oxidoreductase 1. Proc Natl Acad Sci U S A 2003;100:15065-70. [Crossref] [PubMed]


