Promoter methylation of p16 and DAPK genes in brushing, blood, and tissue samples from patients with nasopharyngeal carcinoma: a systematic meta-analysis
Introduction
Nasopharyngeal carcinoma (NPC) is a unique epithelial cancer of the head and neck with an extremely geographical and ethnic distribution. NPC is a prevalent malignancy in Southeast Asia, where it occurs in 20–30 per 100,000 persons every year; it is particularly prevalent in Southern China and ranges from 15 to 50 cases per 100,000 individuals, whereas it is rare in the Western countries, with an annual incidence rate of less than 1 per 100,000 people (1,2). Radiotherapy alone or in combination with chemotherapy has been used as the primary treatment strategy, providing a generally satisfactory disease control in patients with early-stage NPC (3). However, approximately 60–70% of the patients are diagnosed at later stages (III–IV) with loco-regional lymph node metastases (4). Currently, distant metastasis is considered a typical failure pattern, particularly in advanced patients, with poor prognosis (5-7).
Previous studies have shown that certain etiological factors contribute to the initiation and development of NPC, including infection with Epstein-Barr virus, inflammation, dietary and lifestyle factors, and genetics and epigenetic alterations (8-11). Aberrant DNA methylation of tumor suppressor genes (TSGs), such as deleted in liver cancer 1 (DLC1), is a frequent event in NPC (12,13). The human p16 gene, mapped to chromosome 9p21 and consisting of 3 exons and 2 introns, is a cyclin-dependent kinase inhibitor that plays a key role in cell cycle regulation (14,15). Localized on human chromosome 9p34, death-associated protein kinase (DAPK), a serine/threonine kinase, participates in the modulation of a variety of cellular processes, including apoptosis, autophagy, and inflammation (16,17). The loss of p16 and DAPK expression as TSGs through promoter methylation has been shown to be associated with many carcinomas (18-20).
Nonetheless, there are conflicting findings concerning the relationship between p16 promoter methylation and NPC. For example, Challouf et al. reported the absence of significant association between p16 promoter methylation and NPC (21), whereas the results of Nawaz et al. evidenced the presence of a significant association between p16 promoter methylation and NPC (22). Similarly, there are controversial results on DAPK promoter methylation and its relation to NRC occurrence. For instance, Chang et al. established that the DAPK promoter methylation rate in NPC patients was similar to or even lower than that in healthy subjects (23). In contrast, the investigation of Tian et al. revealed that the level of DAPK promoter methylation was significantly higher in NPC cases than in healthy subjects (24). It is important to note that studies with a small sample size may lack strong statistical power (25,26). Therefore, to address these inconsistencies and evaluate the relationship between p16 and DAPK promoter methylation and NPC, basing on several selection criteria, we integrated all eligible publications on this subject. In addition, we also determined whether p16 and DAPK promoter methylation was linked with the clinical stage of NPC.
Methods
Search strategy and selection criteria
We conducted a comprehensive literature search of four online electronic databases, including PubMed, EBSCO, Cochrane Library, and EMBASE, to identify eligible studies published before August 8th, 2016. We used the following combination of keywords and free words: ‘nasopharyngeal cancer OR nasopharyngeal neoplasm OR nasopharyngeal carcinoma OR nasopharyngeal tumor OR NPC’, ‘p16 OR INK4A OR CDKN2A OR cyclin-dependent kinase inhibitor 2A’, ‘DAPK OR death-associated protein kinase OR DAP-kinase’, ‘methylation OR hypermethylation OR promoter methylation OR epigenetic’.
To be included in our meta-analysis, the articles had to meet the following inclusion criteria: (I) the diagnosis of NPC was based on histopathological examination; (II) case-control or cohort design studies published in English; (III) studies provided sufficient information with regard to the methylation rate of p16 or DAPK promoter methylation to calculate the pooled odds ratios (ORs) and corresponding 95% confidence intervals (CIs); (IV) when identical or overlapping data were used in multiple publications, only the most recent paper or the paper with the largest population was included; (V) the sample type was without restriction and included tissue, blood, and brushing samples from patients with NPC. A tumor stage of ≤1 was considered as an early stage, whereas a tumor stage of ≥2 was defined as a later stage.
Data extraction
Two independent reviewers extracted the following data from each available study using selection standards, including the last name of the first author, publication year, country, ethnicity, sample type, the participants of cases and controls, method for detection of methylation, the frequency of p16 or DAPK promoter methylation, p16 or DAPK expression status. Nontumorous specimens were used as control samples. Any disagreements concerning study selection and data extraction were resolved by consensus or by a third reviewer.
Statistical analysis
In the current meta-analysis, statistical analysis of the pooled data was performed using Stata software, version 12.0 (STATA Corp., College Station, TX, USA). The pooled OR and corresponding 95% CI were calculated to determine the strength of the association between p16 or DAPK promoter methylation and NPC risk. In addition, the correlation between p16 or DAPK promoter methylation and the clinical stage of NPC was also established. The statistical heterogeneity among the studies was tested based on the Cochran’s Q and I2 tests (27). The random-effects model was employed when the P value was less than 0.1 in the Q-test, indicating the presence of substantial heterogeneity; otherwise, the fixed-effects model was applied when no evidence of the heterogeneity was observed (28,29). P value <0.05 was considered statistically significant.
Results
Study characteristics
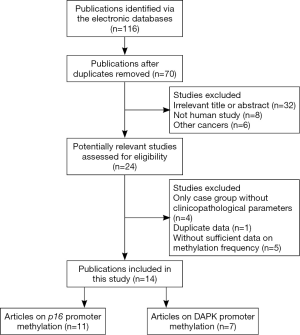
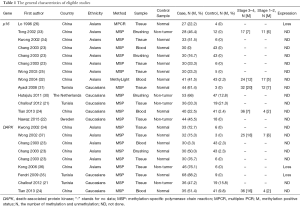
Initially, a total of 116 potential articles were collected by an extensive search of the databases used (Figure 1). After selection based on the eligibility criteria,, eleven available articles on p16 promoter methylation (21-26,30-34) and seven eligible articles on DAPK promoter methylation (21,23,24,34-37) were eventually identified in this analysis. Seven of these studies evaluated the association between p16 promoter methylation and NPC in NPC vs. nontumorous tissues (21-23,25,26,31,34). Three studies evaluated the relationship between p16 promoter methylation and NPC in NPC vs. normal blood samples (23,24,32). In addition, three studies estimated the connection between p16 promoter methylation and NPC in NPC vs. nontumorous brushing samples (23,30,33). Six other studies assessed the correlation between DAPK promoter methylation and NPC in NPC vs. nontumorous tissues (21,23,34-37), and two studies evaluated the association between DAPK promoter methylation and NPC in NPC vs. normal blood samples (23,24). The relationship between DAPK promoter methylation and NPC in NPC vs. normal brushing samples was estimated in one study (23) and the correlation of p16 promoter methylation with tumor stage in NPC in four (24,31-33). Two studies assessed the association of DAPK promoter methylation with tumor stage in NPC (24,37). Table 1 presents the main characteristics of the included studies.
Full table
Correlation of p16 promoter methylation in cancer vs. controls
No obvious heterogeneity was observed for p16 promoter methylation in cancer vs. controls (P value of the heterogeneity >0.1), and the fixed-effects model was applied. A higher frequency of p16 promoter methylation in NPC than in nontumorous tissues (OR =5.49; 95% CI, 2.39–12.63; P<0.001) was established in seven studies with samples from 244 NPC and 61 nontumorous tissues (Figure 2).
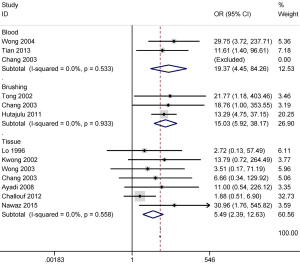
We also evaluated the association between p16 promoter methylation and the risk of NPC in fluid samples (blood and brushing samples). The results from three studies including 111 NPC and 127 normal blood samples demonstrated that p16 promoter methylation in NPC samples was notably higher than in normal blood samples (OR =19.37; 95% CI, 4.45–84.26; P<0.001) (Figure 2). Three other studies with 111 NPC and 102 noncancerous brushing samples evidenced that p16 promoter methylation was significantly higher in NPC samples than in noncancerous brushing samples (OR =15.03; 95% CI, 5.92–38.17; P<0.001) (Figure 2).
Therefore, our results revealed that p16 promoter methylation was significantly correlated with the increased risk of NPC in tissue, blood, and brushing specimens.
Correlation of DAPK promoter methylation in cancer vs. controls
There was no substantial evidence of heterogeneity in cancer vs. controls; thus, the fixed-effects model was used for DAPK promoter methylation (P value of the heterogeneity >0.1). The results concerning six studies with 245 NPC and 49 nontumorous tissue samples indicated that a higher DAPK promoter methylation rate was observed in NPC tissues in comparison to that in nontumorous tissues (OR =17.51; 95% CI, 7.08–43.32; P<0.001) (Figure 3).
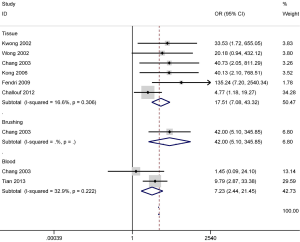
Additionally, the pooled OR of two studies with 65 NPC and 84 normal blood samples showed that DAPK promoter methylation was significantly greater in NPC than in normal blood samples (OR =7.23; 95% CI, 2.44–21.45; P<0.001) (Figure 3). The combined OR in one study exhibited a significant association between DAPK promoter methylation and NPC based on the results from the analysis of 30 NPC and 43 normal brushing samples (OR =42.00; 95% CI, 5.10–345.85; P=0.001) (Figure 3).
Our findings revealed that the methylation status of DAPK promoter was significantly associated with the increased risk of NPC in tissue, blood, and brushing specimens. However, the analyses of brushing and blood samples should be cautiously interpreted as only one or two studies with small sample sizes were involved in this meta-analysis.
Subgroup analysis by ethnic population in cancer vs. nontumorous tissues
Further, we conducted subgroup analysis by ethnicity to evaluate the different degree of association of the stratified population in tissue samples. Subgroup analysis based on ethnicity showed that p16 promoter methylation was significantly correlated with an increased risk of NPC in both among both populations investigated, Asians (OR =5.90; 95% CI, 1.35–25.82; P=0.018) and Caucasians (OR =5.28; 95% CI, 1.93–14.43; P=0.001) (Figure 4).
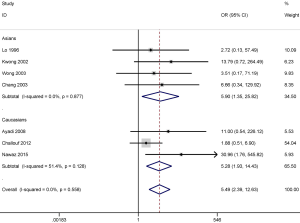
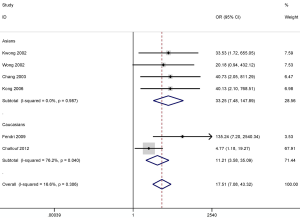
DAPK promoter methylation also was significantly correlated with the increased risk of NPC in the Asian (OR =33.25; 95% CI, 7.48–147.89; P<0.001) and the Caucasian population (OR =11.21; 95% CI, 3.58–35.09; P<0.001) (Figure 5). However, the results of subgroup analysis should be carefully considered as only small sample sizes were included in this study, especially in the Caucasian population subgroup.
Correlation of p16 or DAPK promoter methylation with tumor stage in NPC
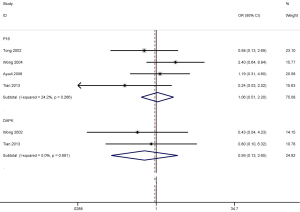
We also determined whether p16 or DAPK promoter methylation was correlated with tumor stage in NPC using the fixed-effects model (Figure 6). The overall OR from four studies involving 109 advanced NPC patients and 44 early NPC patients demonstrated that p16 promoter methylation was not correlated with tumor stage (OR =1.06; 95% CI, 0.51–2.20; P=0.876). The overall OR from two other studies including 61 advanced NPC patients and 11 early NPC patients indicated that DAPK promoter methylation was not associated with tumor stage (OR =0.59; 95% CI, 0.13–2.65; P=0.491). Nonetheless, the results regarding the relationship between p16 or DAPK promoter methylation and tumor stage should be cautiously interpreted as only a small sample size of NPC patients were analyzed in the current research.
Discussion
The hypermethylation of TSGs and hypomethylation of oncogenes are two important molecular mechanisms in epigenomic regulation that play key roles in the initiation and progression of cancer (38-40). The promoter methylation of TSGs may affect cell proliferation, cell death, cell migration, and cell invasion (41). Aberrant promoter methylation of p16 and DAPK genes has been reported in NPC (21,23,34). As a key cell cycle regulator, the p16 gene is involved in the inhibition of cell cycle progression, and the restoration of its expression may induce G0/G1 arrest and suppress the tumorigenic growth of NPC cells (42). A correlation was found between p16 promoter methylation and its expression in NPC, with the absence of p16 expression (26). Earlier studies showed that the loss of DAPK expression was associated with its promoter methylation in NPC (35,36). The inactivation of p16 and DAPK through promoter methylation may play a key in NPC tumorigenesis (43).
However, there are still inconsistent and controversial results regarding the methylation rate of p16 and DAPK promoter in NPC specimens. For example, different promoter methylation rates of the p16 gene, ranging from 22.2% (26) to 61.4% (31), were established in NPC tissues. Inconsistent findings on the frequency of DAPK promoter methylation in NPC tissues, which ranged from 47.2% (21) to 88.2% (35) were also reported previously. Therefore, we systematically investigated studies of p16 and DAPK promoter methylation in NPC samples to estimate the association between p16 and DAPK promoter methylation and NPC.
p16 and DAPK promoter methylation rates were shown to be significantly higher in NPC than in nontumorous tissue samples from the nasopharynx, suggesting that p16 or DAPK promoter methylation may play a pivotal role in the tumorigenesis of NPC. No significant heterogeneity was found in our study, indicating the stability of our results.
The subgroup analysis by ethnic population comparing p16 and DAPK promoter methylation in NPC and nontumorous tissues from the nasopharynx revealed that the promoter methylation of p16 or DAPK was significantly correlated with the risk of NPC in both the Asian and Caucasian populations, indicating that p16 and DAPK promoter methylation may be susceptible genes for Asian and Caucasian populations. In addition, the Asian population had a higher OR of DAPK promoter methylation than the Caucasian population subgroup (33.25 vs. 11.21), which suggested that the Asian population may be more susceptible to DAPK promoter methylation. However, due to the limitation of the small sample size, the analysis of the Caucasian population subgroup in the current study should be cautiously interpreted.
Chang et al. reported that the promoter methylation rate of p16 was 0% in NPC blood samples and 16.7% in NPC brushing samples, respectively (23). On the other hand, Wong et al. reported that a p16 promoter methylation rate of 41.5% detected in NPC blood samples (32), and Hutajulu et al. discovered that p16 promoter had a methylation frequency of 66% in NPC brushing samples (30). Detection of DNA methylation in brushing or blood samples, shows a great potential to be applied as an invasive biomarker for early detection of NPC (44). Previous studies suggest that p16 and DAPK promoter methylation identified in brushing or blood samples may become a useful biomarker in NPC (24,33). Our findings demonstrate that p16 promoter methylation established in blood and brushing samples is significantly associated with the risk of NPC and its rate is significantly higher in blood or brushing samples of NPC patients than in those of healthy subjects, suggesting that p16 promoter methylation may be used as a noninvasive biomarker for NPC detection in blood and brushing samples.
Chang et al. discovered that DAPK promoter methylation frequency was 3.3% in NPC blood samples (23), but a much higher value (51.4%) was reported by Tian et al. (24). On the other hand, a frequency of DAPK promoter methylation in NPC brushing samples that reaches 50% has been reported only in the study of Chang et al. (23). Our results indicated that DAPK promoter methylation in blood and brushing samples was significantly correlated with the risk of NPC and its level was significantly higher in the blood or brushing samples of NPC patients than in those of healthy subjects, indicating that DAPK promoter methylation is a potential noninvasive biomarker for the detection of NPC in blood or brushing samples. Additionally, Tian et al. evidenced that the combined P16 and DAPK promoter methylation did not significantly increase the potential capacity for NPC detection, but the combination of four-gene marker can be applied as a promising tool for the diagnosis of NPC (24). However, more clinical research studies are required to further validate these findings in the future. Only the OR of one study with brushing samples was included in this study. Thus, we should carefully consider the results from the analysis of blood and brushing samples conducted for the detection of p16 and DAPK promoter methylation. Additional studies with larger sample sizes are needed to confirm our results.
Finally, we also investigated whether p16 or DAPK promoter methylation was correlated with the clinical stage of NCR and found that p16 and DAPK promoter methylation was not associated with tumor stage. More studies are exceedingly essential to validate that finding in the future.
Some limitations of this meta-analysis should be acknowledged. First, only articles published in English were included in this research. Articles in other languages and publications of other types, such as conferences abstracts, were excluded due to their unreadable contents or insufficient information, which might have led to a selection bias. Second, due to the limitation of insufficient data, we did not assess the relationship between p16 and DAPK promoter methylation and other clinicopathological features, such as tumor grade, sex status, and lymph node status. Third, further studies with larger sample sizes should be done to validate our results, especially those concerning DAPK promoter methylation in blood and brushing specimens.
In conclusion, our findings suggest that p16 and DAPK promoter methylation may play an important role in NPC development. The promoter methylation levels of p16 or DAPK are potential useful biomarkers for NPC detection in blood and brushing samples in clinical settings. No significant association was found between p16 or DAPK promoter methylation and tumor stage. Due to the limitations of the sample size in the present analysis, further large-scale studies with larger sample sizes of subjects are necessary to investigate more comprehensively the clinical effects of p16 and DAPK promoter methylation in NPC patients.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.12.08). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wu JM, Yu TJ, Yeh SA, et al. Use dose bricks concept to implement nasopharyngeal carcinoma treatment planning. Biomed Res Int 2014;2014:720876.
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [Crossref] [PubMed]
- Yi JL, Gao L, Huang XD, et al. Nasopharyngeal carcinoma treated by radical radiotherapy alone: Ten-year experience of a single institution. Int J Radiat Oncol Biol Phys 2006;65:161-8. [Crossref] [PubMed]
- Mao YP, Xie FY, Liu LZ, et al. Re-evaluation of 6th edition of AJCC staging system for nasopharyngeal carcinoma and proposed improvement based on magnetic resonance imaging. Int J Radiat Oncol Biol Phys 2009;73:1326-34.
- Chen L, Hu CS, Chen XZ, et al. Concurrent chemoradiotherapy plus adjuvant chemotherapy versus concurrent chemoradiotherapy alone in patients with locoregionally advanced nasopharyngeal carcinoma: a phase 3 multicentre randomised controlled trial. Lancet Oncol 2012;13:163-71. [Crossref] [PubMed]
- Ma BB, Chan AT. Systemic treatment strategies and therapeutic monitoring for advanced nasopharyngeal carcinoma. Expert Rev Anticancer Ther 2006;6:383-94. [Crossref] [PubMed]
- Liu MT, Hsieh CY, Chang TH, et al. Prognostic factors affecting the outcome of nasopharyngeal carcinoma. Jpn J Clin Oncol 2003;33:501-8. [Crossref] [PubMed]
- Zhang Y, Zhou GQ, Liu X, et al. Exploration and Validation of C-Reactive Protein/Albumin Ratio as a Novel Inflammation-Based Prognostic Marker in Nasopharyngeal Carcinoma. J Cancer 2016;7:1406-12. [Crossref] [PubMed]
- Tsao SW, Yip YL, Tsang CM, et al. Etiological factors of nasopharyngeal carcinoma. Oral Oncol 2014;50:330-8. [Crossref] [PubMed]
- Xue WQ, Qin HD, Ruan HL, et al. Quantitative association of tobacco smoking with the risk of nasopharyngeal carcinoma: a comprehensive meta-analysis of studies conducted between 1979 and 2011. Am J Epidemiol 2013;178:325-38. [Crossref] [PubMed]
- Lo KW, To KF, Huang DP. Focus on nasopharyngeal carcinoma. Cancer Cell 2004;5:423-8. [Crossref] [PubMed]
- Bruce JP, Yip K, Bratman SV, et al. Nasopharyngeal Cancer: Molecular Landscape. J Clin Oncol 2015;33:3346-55. [Crossref] [PubMed]
- Feng X, Ren C, Zhou W, et al. Promoter hypermethylation along with LOH, but not mutation, contributes to inactivation of DLC-1 in nasopharyngeal carcinoma. Mol Carcinog 2014;53:858-70. [Crossref] [PubMed]
- Piepkorn M. Melanoma genetics: an update with focus on the CDKN2A(p16)/ARF tumor suppressors. J Am Acad Dermatol 2000;42:705-22; quiz 23-6. [Crossref] [PubMed]
- Lukas J, Parry D, Aagaard L, et al. Retinoblastoma-protein-dependent cell-cycle inhibition by the tumour suppressor p16. Nature 1995;375:503-6. [Crossref] [PubMed]
- Huang Y, Chen L, Guo L, et al. Evaluating DAPK as a therapeutic target. Apoptosis 2014;19:371-86. [Crossref] [PubMed]
- Cohen O, Kimchi A. DAP-kinase: from functional gene cloning to establishment of its role in apoptosis and cancer. Cell Death Differ 2001;8:6-15. [Crossref] [PubMed]
- Misawa K, Mochizuki D, Imai A, et al. Prognostic value of aberrant promoter hypermethylation of tumor-related genes in early-stage head and neck cancer. Oncotarget 2016;7:26087-98. [PubMed]
- Laskar RS, Ghosh SK, Talukdar FR. Rectal cancer profiling identifies distinct subtypes in India based on age at onset, genetic, epigenetic and clinicopathological characteristics. Mol Carcinog 2015;54:1786-95. [Crossref] [PubMed]
- Brait M, Loyo M, Rosenbaum E, et al. Correlation between BRAF mutation and promoter methylation of TIMP3, RARbeta2 and RASSF1A in thyroid cancer. Epigenetics 2012;7:710-9. [Crossref] [PubMed]
- Challouf S, Ziadi S, Zaghdoudi R, et al. Patterns of aberrant DNA hypermethylation in nasopharyngeal carcinoma in Tunisian patients. Clin Chim Acta 2012;413:795-802. [Crossref] [PubMed]
- Nawaz I, Moumad K, Martorelli D, et al. Detection of nasopharyngeal carcinoma in Morocco (North Africa) using a multiplex methylation-specific PCR biomarker assay. Clin Epigenetics 2015;7:89. [Crossref] [PubMed]
- Chang HW, Chan A, Kwong DL, et al. Evaluation of hypermethylated tumor suppressor genes as tumor markers in mouth and throat rinsing fluid, nasopharyngeal swab and peripheral blood of nasopharygeal carcinoma patient. Int J Cancer 2003;105:851-5. [Crossref] [PubMed]
- Tian F, Yip SP, Kwong DL, et al. Promoter hypermethylation of tumor suppressor genes in serum as potential biomarker for the diagnosis of nasopharyngeal carcinoma. Cancer Epidemiol 2013;37:708-13. [Crossref] [PubMed]
- Wong TS, Tang KC, Kwong DL, et al. Differential gene methylation in undifferentiated nasopharyngeal carcinoma. Int J Oncol 2003;22:869-74. [PubMed]
- Lo KW, Cheung ST, Leung SF, et al. Hypermethylation of the p16 gene in nasopharyngeal carcinoma. Cancer Res 1996;56:2721-5. [PubMed]
- Coory MD. Comment on: Heterogeneity in meta-analysis should be expected and appropriately quantified. Int J Epidemiol 2010;39:932; author reply 933.28.
- Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ 2003;327:557-60. [Crossref] [PubMed]
- DerSimonian R. Meta-analysis in the design and monitoring of clinical trials. Stat Med 1996;15:1237-48; discussion 49-52. [Crossref] [PubMed]
- Hutajulu SH, Indrasari SR, Indrawati LP, et al. Epigenetic markers for early detection of nasopharyngeal carcinoma in a high risk population. Mol Cancer 2011;10:48. [Crossref] [PubMed]
- Ayadi W, Karray-Hakim H, Khabir A, et al. Aberrant methylation of p16, DLEC1, BLU and E-cadherin gene promoters in nasopharyngeal carcinoma biopsies from Tunisian patients. Anticancer Res 2008;28:2161-7. [PubMed]
- Wong TS, Kwong DL, Sham JS, et al. Quantitative plasma hypermethylated DNA markers of undifferentiated nasopharyngeal carcinoma. Clin Cancer Res 2004;10:2401-6. [Crossref] [PubMed]
- Tong JH, Tsang RK, Lo KW, et al. Quantitative Epstein-Barr virus DNA analysis and detection of gene promoter hypermethylation in nasopharyngeal (NP) brushing samples from patients with NP carcinoma. Clin Cancer Res 2002;8:2612-9. [PubMed]
- Kwong J, Lo KW, To KF, et al. Promoter hypermethylation of multiple genes in nasopharyngeal carcinoma. Clin Cancer Res 2002;8:131-7. [PubMed]
- Fendri A, Masmoudi A, Khabir A, et al. Inactivation of RASSF1A, RARbeta2 and DAP-kinase by promoter methylation correlates with lymph node metastasis in nasopharyngeal carcinoma. Cancer Biol Ther 2009;8:444-51. [Crossref] [PubMed]
- Kong WJ, Zhang S, Guo CK, et al. Effect of methylation-associated silencing of the death-associated protein kinase gene on nasopharyngeal carcinoma. Anticancer Drugs 2006;17:251-9. [Crossref] [PubMed]
- Wong TS, Chang HW, Tang KC, et al. High frequency of promoter hypermethylation of the death-associated protein-kinase gene in nasopharyngeal carcinoma and its detection in the peripheral blood of patients. Clin Cancer Res 2002;8:433-7. [PubMed]
- Franco R, Schoneveld O, Georgakilas AG, et al. Oxidative stress, DNA methylation and carcinogenesis. Cancer Lett 2008;266:6-11. [Crossref] [PubMed]
- Corson TW, Gallie BL. One hit, two hits, three hits, more? Genomic changes in the development of retinoblastoma. Genes Chromosomes Cancer 2007;46:617-34. [Crossref] [PubMed]
- Bodmer WF. 1998 Runme Shaw Memorial Lecture: somatic evolution of cancer. Ann Acad Med Singapore 1999;28:323-9. [PubMed]
- Maziveyi M, Alahari SK. Breast Cancer Tumor Suppressors: A Special Emphasis on Novel Protein Nischarin. Cancer Res 2015;75:4252-9. [Crossref] [PubMed]
- Wang GL, Lo KW, Tsang KS, et al. Inhibiting tumorigenic potential by restoration of p16 in nasopharyngeal carcinoma. Br J Cancer 1999;81:1122-6. [Crossref] [PubMed]
- Lo KW, Chung GT, To KF. Deciphering the molecular genetic basis of NPC through molecular, cytogenetic, and epigenetic approaches. Semin Cancer Biol 2012;22:79-86. [Crossref] [PubMed]
- Yang X, Dai W, Kwong DL, et al. Epigenetic markers for noninvasive early detection of nasopharyngeal carcinoma by methylation-sensitive high resolution melting. Int J Cancer 2015;136:E127-35. [Crossref] [PubMed]