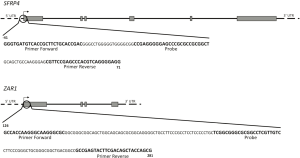
Evaluation of DNA methylation in promoter regions of SFRP4 and ZAR1 in urine and plasma of women with cervical lesions
Introduction
Cervical cancer (CC) is the third most common cancer in women of all ages worldwide and the second most common in women between 15 and 44 years old (1). About 500,000 new cases of CC are detected annually, of which about half will die from this disease. The most affected population in the world is found in developing countries, with 84% of new cases and 87% of CC-related deaths (1). In Chile, CC is the sixth cause of death by malignant tumors in women, with a mortality rate of 6 deaths per 100,000 (2).
Preneoplastic lesions of the cervix can be classified as type I (mild dysplasia), type II (moderate dysplasia) and type III (severe dysplasia and carcinoma in situ) cervical intraepithelial neoplasia (CIN) (3-5). These lesions can be also classified according to Papanicolaou test results into the low-grade squamous intraepithelial lesion (L-SIL), which consists of CIN I and/or human papillomavirus (HPV)-infected tissue, and the high-grade squamous intraepithelial lesion (H-SIL) comprising both CIN II and CIN III (5).
The persistent infection with high-risk oncogenic HPV (HR-HPV) is the main etiological cause of CC, being found in 99.7% of the cases (6). However, the presence of a persistent infection for HR-HPV is not enough to transform the epithelial cells of the host. Several other modifications should be present to ultimately trigger immortalization in epithelial cells and induce a malignant and invasive phenotype. This process can involve genetic bases (mutations, deletions, copy-number alterations and chromosomal rearrangements) or epigenetic bases (DNA methylation, post-translational modifications of histones or microRNAs) (7-9).
The most frequently studied epigenetic phenomenon is DNA methylation, defined as the addition of a methyl group to the 5’ carbon of the pyrimidine ring of a cytosine. The abnormal increase of methylation in the promoter regions of tumor suppressor genes (TSG) is known as aberrant hypermethylation, which is a common alteration in carcinogenesis as this could induce a partial or complete repression of the affected genes. Aberrant hypermethylation can be detected in the early stages of cervical carcinogenesis, and it may therefore constitute a promising and useful tool as a biomarker for early detection, progression, survival prognosis and/or therapeutic response in this disease (10-13).
In CC, aberrant hypermethylation affects several TSGs belonging to different pathways involved in cell adhesion, DNA repair and cell cycle control, and also to those genes related to nuclear receptors (14).
In recent years, several studies have been performed to develop a reliable biomarker that can detect the early stages of CC in samples that are less invasive than a cervical brush and as easy to obtain as urine and blood. The detection of DNA hypermethylation in urine has been previously used in bladder, prostate and cervical cancer (15-22) and possesses the advantage that it can be self-collected by the patient. The detection of DNA methylation in blood has been used in several types of malignancies, such as lung (23), colorectal (24), breast (25), and cervical cancer (26-29), among others. The determination of methylation patterns in some specific genes could offer a new alternative to current CC screening programs.
In a previous study, we constructed methylation microarrays (MeDIP-chip) to find highly methylated genes in cancer but not in normal samples to be assessed as potential methylation biomarkers for diagnosis. Data analysis of this MeDIP-chip performed with DNA extracted from the cytobrush samples of 7 patients with cervical squamous cell carcinoma (SCC) and 12 women with normal epithelium showed that there was promoter aberrant hypermethylation of ZAR1 and SFRP4 genes (30). Additionally, our group evaluated the methylation status of these genes in cytobrush samples within different histopathological subgroups of diagnosis (non-lesion: 60; L-SIL: 40; H-SIL: 40 and SCC: 31) through quantitative methylation-specific PCR (QMSP) using specific probes for each gene. Significant differences in the methylation levels between the SCC group and the other groups (P<0.01) were found. The technique sensitivity was 77.4% and 71%; the specificity was 80% and 65%, and the area under the curve (AUC) was up to 0.83 and 0.75 for ZAR1 and SFRP4, respectively (31).
With this background, this study focused on assessing the methylation levels in the promoter regions of ZAR1 and SFRP4, using urine and plasma samples to propose these patterns as potential biomarkers for early detection and/or progression of CC in non-invasive specimens.
Methods
Clinical samples
Samples were obtained between 2011 and 2013 from the Cervical Pathology Healthcare in the Hernán Henríquez Aravena Hospital in Temuco, Chile. A total of 171 paired samples of urine and plasma were taken from women with different histopathological diagnoses: 60 without lesion (non-lesions), 40 L-SIL, 40 H-SIL and 31 SCC. Women who participated in the study signed an informed consent approved by the School of Medicine Ethics Committee of Universidad de La Frontera (Approval No. 246/006). Pathology Unit from Hernán Henríquez Aravena Hospital confirmed the histopathological diagnosis.
Random urine samples (first-catch) were collected in sterile 50-mL flasks containing 3 mL of crystal violet as preserver. The minimum acceptable volume of urine sample was 10 mL. Briefly, these samples were centrifuged at 3,500 rpm for 12 minutes at room temperature (RT) and supernatant was discarded. Then, 500 µL of PBS was added and this mix was taken to a 1.5-mL tube and was centrifuged at 12,000 rpm for 5 min at RT. Supernatant was again discarded and 500 µL of lysis buffer was added to be finally stored at −20 °C.
Blood samples were collected in sterile 5-mL tubes containing EDTA as anticoagulant and DNA preserver. The minimum acceptable volume of blood sample was 4 mL. These samples were centrifuged at 3,500 rpm for 10 minutes at RT and plasma was then taken to 1.5-mL tubes for storage at −80 °C.
DNA extraction
DNA from urine and plasma samples was extracted using the EZNA Tissue DNA Extraction kit (Omega, USA), according to the manufacturer’s instructions. DNA quantity and purity was evaluated with a Nanodrop-1000 spectrophotometer. Only samples with a DNA concentration over 20 ng/µL and purity over 1.7 according to an A260/A280 coefficient were used. DNA integrity was evaluated by PCR amplification of a 268-base pair fragment of β-globin (HBB) using the following primers PCO4 (5' CAA CTT CAT CCA CGT TCA CC 3') and GH20 (5' GAA GAG CCA AGG ACA GGT AC 3') (32).
Bisulfite modification
1 µg of genomic DNA was modified using the EZ DNA Methylation Kit-GoldTM (Zymo Research, USA) according to the manufacturer’s instructions. Bisulfite modification was confirmed by the amplification of a 133-bp fragment of β-actin (ACTB) (Table 1).

Full table
Quantitative methylation-specific PCR (QMSP)
This qPCR-based method that measures fluorescent emission was performed to determine methylation levels of the promoter regions of SFRP4 and ZAR1. Primers and probe sequences of SFRP4 and ZAR1 are shown in Table 1 and the sequences to which these primers and probes bind are shown in Figure 1. Amplification reactions were made in triplicate in a final volume of 20 µL with 1 µL of bisulfite-modified DNA; 300 nM of each primer; 50 mM of probe; 0.375 units of Platinum Taq Polymerase (Invitrogen, USA); 100 µL of dNTPs; 100 nM of ROX dye (Invitrogen, USA), 8.3 mM of ammonium sulfate; 33.5 mM of Trizma (Sigma, USA); 3.35 mM of magnesium chloride; 5 mM of 2-mercaptoethanol; and 0.05% of DMSO. Amplification was performed according to the following thermic profile: 95 °C for 10 minutes, followed by 40 cycles of 95 °C for 30 seconds, 56 °C for 1 minute and 72 °C for 30 seconds, using Mx3000P QPCR equipment (Stratagene, USA). Each PCR reaction included bisulfite-modified DNA samples; a 100% methylated DNA (Zymo Research, USA) as positive control, leukocyte DNA from a healthy person as negative control and, finally, several blanks of PCR mix without DNA. Serial dilutions (250, 50, 10, 5 and 2 ng) of positive control (Universal Methylated Human DNA Standard, Zymo Research) were used for standard curve construction in order to obtain equation of the line and slope values for each gene, which served subsequently to calculate the amount of methylated DNA of each reaction. Relative DNA methylation levels for SFRP4 and ZAR1 were determined as the relation between the specific methylation of the amplified gene and ACTB (reference gene), multiplied by 1,000 for easier tabulation (mean value of triplicates of study gene, divided by mean value of triplicates of ACTB, multiplied by 1,000).
ROC analysis
Receiver operational curves (ROCs) were calculated using relative DNA methylation levels obtained by QMSP comparing normal women and CC patients for each gene (SFRP4 and ZAR1) and each sample type (urine and plasma). The AUC was calculated to compare different diagnostic techniques. According to Swets et al. (33), an AUC ≥0.70 is moderately good for group discrimination (negative vs. cancer).
Statistical analysis
A chi-squared test was performed to compare ages and histological diagnoses, and Fisher’s exact test for group comparison. A Kruskal Wallis test and Dunn’s post-test were used to compare DNA methylation levels among groups. A 95% confidence was used for each test. Analyses and graphs were made using Prism GraphPad 5.0 (GraphPad Software, Inc., USA). Cohen’s kappa coefficient was calculated for concordance analysis of the results of the different sample types. The positivity of the tests was compared according to the cut-off point obtained with the ROC curves. Another comparison was made using the results of the presence or absence of methylation. Each analysis was performed using SPSS v.20.0 (SPSS IBM Inc., USA). Finally, the percentage of coincidence in methylation status among the different sample types (urine, plasma and the previous cytobrush results) was calculated, multiplying by 100 the number of methylated samples in urine or plasma and dividing this result by the total number of methylated samples in cytobrush or urine samples (these samples showed the highest number of positive results) according to each case.
Results
Sample characteristics
The age range of participants was between 16 and 81 years old (mean: 36.4 years). The samples were grouped according to the age in ≤35 (57.3%) and >35 (42.7%) years. There was an association between age and grade of cervical lesion (P<0.0001, Chi square, 95% confidence), with a significant difference being observed between the CC patient group and the other groups (non-lesion P=0.0004; L-SIL and H-SIL P<0.0001, Fisher’s exact test, 95% confidence).
Most patients without lesions and those with L-SIL and H-SIL were ≤35 years, whereas most patients with CC were >35 years (Table 2). Eight samples of preneoplastic lesions did not have an accurate diagnosis and were discarded (four L-SIL and four H-SIL). Three plasma samples were discarded because the ACTB gene could not be amplified after its bisulfite modification, even after DNA re-extraction from the samples. All the other samples had an adequate DNA integrity (HBB positive) and were properly converted by bisulfite modification (ACTB positive).

Full table
DNA methylation in urine samples
The methylation levels in promoter regions of the SFRP4 and ZAR1 genes were evaluated by QMSP in 171 urine samples (non-lesions: 60, L-SIL: 40, H-SIL: 40 and SCC: 31). For SFRP4 showed significant differences in patients with SCC (mean =4.98) compared to all the other groups: non-lesion (mean =0; P<0.0001), L-SIL (mean =0.04098; P=0.0007) and H-SIL (mean =0.07513; P=0.0034) (Figure 2A). However, for ZAR1 no differences were observed between non-lesion (mean: 6.155), L-SIL (mean =79.48), H-SIL (mean =13.71) and SCC groups (mean =1.424) (Figure 2B).
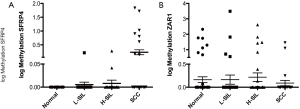
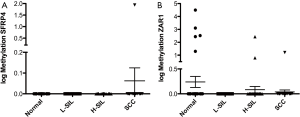
DNA methylation in plasma samples
Methylation levels in promoter regions of SFRP4 and ZAR1 were evaluated by QMSP in 168 plasma samples (non-lesion: 58, L-SIL: 40, H-SIL: 39 and SCC: 31). There was no significant difference in DNA methylation levels either for SFRP4 or ZAR1 among the study groups (Figure 3A,B).
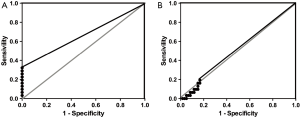
ROC curves for QMSP in urine and plasma samples
The ROC analysis results of DNA methylation in urine showed an AUC of 0.6613 and 0.5065 for SFRP4 and ZAR1, respectively. The best cut-off point for SFRP4 was 0.03026 with a specificity of 100% and a sensitivity of 30.26% (Figure 4A, Table 3). For ZAR1 the best cut-off point was 0.09571 with a specificity of 83.33% and a sensitivity of 19.35% (Figure 4B, Table 3). When the methylation in both genes was evaluated simultaneously, an AUC of 0.633 was obtained, with a specificity of 83.3% and a sensitivity of 45.16%, and the best cut-off point was 0.03026 (Table 3).

Full table
In plasma samples an AUC of 0.5272 and 0.5284 was obtained for SFRP4 and ZAR1, respectively. The best cut-off point for SFRP4 was 43.33, with a sensitivity of 98.28% and a specificity of 3.226% (Table 3). For ZAR1 the best cut-off point was 8.083, with a specificity of 96.77% and a sensitivity of 8.621% (Table 3). An AUC of 0.6333 was obtained with a sensitivity of 93.55% and a specificity of 8.3% when both genes were analyzed (Table 3). ROC curves for methylation results in plasma are not shown because the curves were close to the identity line (AUC =0.5), which did not offer more information.
Concordance analysis
For this analysis, the methylation status of SFRP4 and ZAR1 in urine and plasma was compared to methylation results of these two genes obtained in cytobrush samples, which were previously reported (31).
Presence or absence of DNA methylation
In order to analyze the presence or absence of methylation in promoter region of SFRP4 among the different types of samples, Cohen’s Kappa coefficient was calculated, obtaining values of 0.072; −0,047 and 0.085 when results from urine vs. cytobrush, plasma vs. cytobrush, and plasma vs. urine were compared, respectively. In the case of ZAR1 these values were 0.055; −0.022 and 0.041, respectively (Table 4).

Full table
Positivity according to the calculated cut-off point
When positive or negative results were analyzed according to the calculated cut-off for SFRP4, Cohen’s kappa coefficient was 0.80; −0.035 and 0.099 for urine vs. cytobrush, plasma vs. cytobrush and plasma vs. urine, respectively. The values obtained for ZAR1 were 0.055; −0.011 and 0.052, respectively (Table 4).
Coincidence of results
For this analysis, the methylation status of SFRP4 and ZAR1 in urine and plasma was also compared their methylation status in cytobrush samples (31).
Presence or absence of DNA methylation
The coincidence percentages among the different types of paired samples were assessed using the above-mentioned results of presence or absence of methylation in the promoter region of SFRP4. The coincidence percentages were 69.2%, 0% and 25% for urine vs. cytobrush, plasma vs. cytobrush and plasma vs. urine, respectively. For ZAR1 these values were 75%, 50% and 25%, respectively (Table 4).
Positivity according to the calculated cut-off point
When positive or negative results were analyzed according to the calculated cut-off considering the coincidence percentage for SFRP4, these values were 69.2%, 0% and 25% for urine vs. cytobrush, plasma vs. cytobrush and plasma vs. urine, respectively. The coincidence percentages for ZAR1 were 75%, 57% and 28%, respectively (Table 4).
Discussion
CC has among the highest prevalence and incidence in women worldwide, especially in developing countries (34). The main screening technique for this malignancy—or its precursor lesions—is the Papanicolaou stain (PAP), which, despite having greatly reduced the prevalence of CC, has a lower sensitivity than 60%. Therefore, in recent years new HPV-based tests have emerged showing better sensitivity than the PAP smear in CC diagnoses. Nevertheless, these tests cannot predict which patients will progress to invasive cancer.
Several reports have found that promoter hypermethylation of certain genes is an early event in cancer development (35), given that it regulates gene expression. In CC, aberrant methylation in genes such as p16INK4A (36), CDH1 (7), hMLH1, VHL, APC (37), SFRP4, ZAR1 (31), FKBP6 and ZNFS16 (30) have been proposed as prognostic and/or progression markers for CC in cervical scrapes. However, cytobrush sampling can be affected in a population by factors such as shame, religion or sociocultural background, and so forth. Therefore, the use of other samples such as blood and urine that would allow increased coverage of CC screening programs are worthy of assessment. Some advantages of urine samples compared to cervical samples are that sampling is more acceptable to women (38), urine does not interfere with the natural history of HPV infection (39), and higher concentrations of genomic DNA can be detected (83–100%) (40), which supports its use for hypermethylated gene detection.
In this study a significant difference was found in the methylation levels of SFRP4 among samples from patients with SCC and other groups of women, which is supported by the results obtained for this gene in cytobrush samples (31). However, no difference in methylation levels of ZAR1 was detected in urine and plasma among the study groups.
There was a marked decrease in sensitivity for the discrimination between women with normal epithelium or cancer, comparing cytobrush samples (77.4% ZAR1; 71% SFRP4) and urine (19.35% ZAR1; 30.26% SFRP4). AUC values showed a decrease from 0.7476 to 0.6613 in the case of SFRP4 and from 0.8296 to 0.5065 for ZAR1, which were termed as less accurate according to Swets et al.’s classification (AUC between 0.5–0.7) (31,33). Feng et al. obtained similar sensitivities when they studied individual genes (between 6% and 47% for CDH13, RARB, DAPK1 and TWIST genes). On the other hand, sensitivity increased to 45.61% when the methylation levels of the two genes (SFRP4 and ZAR1) were combined, which was also similar to the results reported by Feng et al. (45%) (22). In addition, these sensitivities were similar to the results obtained with cytology test of cervical exfoliated cells (30% to 60%) (22).
Higher sensitivities were obtained by Chung et al. in bladder cancer (sensitivity of 85% using a panel of five genes) (19), and by Rouprêt et al. in prostate cancer (sensitivity of 86% using a panel of four genes) (41), that could be explained by the anatomical path of urine elimination.
Some of the main difficulties that may interfere with DNA methylation detection in urine are: (I) in these cases, samples are not collected from the original site of disease, and only contain spontaneously exfoliated cells (39) or certain amounts of trans-renal DNA from patients or from pathogens (42-44); (II) cells of interest are more diluted in urine sample; (III) urine contains PCR inhibitors (45); and (IV) human DNA levels in urine are not constant during evacuation, having higher concentrations in the early fraction than in the medium fraction or total urine (46). Therefore, the first fraction of urine should be used in future studies to obtain a higher number of exfoliated cervical cells and thus a larger amount of DNA. Also, using biomarkers panels or establishing methylation scores (algorithms) could increase the detection sensitivity and turn urine into a useful sample for detecting CC or other types of cancer.
Detection of DNA methylation in plasma samples
Several studies have found that aberrant methylation of specific genes can be detected in DNA extracted from plasma or serum from patients with various malignancies (23-28,47,48), which has a particular clinical benefit for achieving an early molecular diagnosis and assessing progression in these diseases (49-51).
In this study, no differences in DNA methylation levels were found among the different women’s groups for either of the two study genes. This result is due to the low methylation frequency observed in the cancer group for ZAR1 and SFRP4 in plasma samples (presence in 1/31 samples or 3.22%). These frequencies are similar to those obtained by Widschwendter et al. for the hTERT and TIMP3 genes (no more than 4%) in CC (26). However, higher frequencies have been found by studying the methylation status of the CALCA (62%), PGR (79%) (26) and DAPK genes (64.3%) (27) in patients with CC.
The lower methylation frequency of these genes could be explained by the way CC is propagated. The main dissemination route is local extension and lymphatic embolization. However, patients with larger lesions or with a more advanced disease can have a hematogenous spread by direct invasion of blood vessels through capillaries and lacerated veins, through the thorax or through small venous and lymphatic channels (52). CC usually follows the path of least resistance such as lateral spread, which involves the parametrium. This could explain why only a few cells or free DNA molecules are introduced into the blood stream, hindering the detection of methylated DNA in plasma samples. A similar phenomenon occurs in the case of urine because a smaller number of DNA and CC cells can cross the renal filtration. Another possibility is that the tumors are small or present less progression.
There are some nucleases in the blood stream that can degrade cell-free DNA (cfDNA). The cfDNA present in the blood is cleared by the liver and kidney, having a variable half-life in circulating blood ranging from 15 minutes to several hours (53).
To date, the correlational evidence between DNA methylation observed in cancer tissues compared to blood is limited. In different nutritional interventions, DNA methylation measurements from blood do not always reflect the methylation status of other tissues (54). More research is needed to understand the correlation between methylation patterns of a specific tissue and other sample types.
Another approach that could be considered for the development of biomarkers is the use of the DNA contained in exosomes. Exosomes are small cell-derived vesicles with a diameter of between 50 and 150 nm composed of a lipid bilayer containing membrane proteins. These have been suggested as active transporters for proteins, lipids and nucleic acids including microRNAs (miRNAs), mRNA, DNA, lncRNAs (long non-coding RNA), and other non-coding RNAs, that can be protected from enzymatic degradation (55). As exosomes have been found in several sample types such as urine, serum, plasma and cervical vaginal lavages (56), their use as biomarkers for cancer detection is promising.
One of the major limitations of this study was the fact that fractionation and selection of the exosomes or circulating tumor cells (CTCs) was not considered in the pre-analytical steps of these experiments. These pre-analytical steps could have helped enhance the amount and quality of DNA obtained from samples and, subsequently, the results could have shown a higher methylation frequency of the ZAR1 and SFRP4 genes in these samples in order to establish a stronger correlation with previously reported data.
Concordance analysis and coincidence between different types of samples
These analyses compared methylation status of SFRP4 and ZAR1 in urine and plasma to the methylation status of SFRP4 and ZAR1 in cytobrush samples, reported by previously Brebi et al. (31). Cohen’s kappa coefficients showed low correlations between results of methylation presence and positivity for each study gene through the different sample types (k<0.4) (57). These values are given mainly by the low detection of methylation in urine samples and the lowest detection in plasma samples. However, comparing the coincidence percentage of urine versus cytobrush samples a higher correlation was found for both genes. Conversely, this coincidence was not seen when plasma was compared with other sample types. The higher coincidence percentage of cytobrush with urine and not with plasma can be explained for the greater amount of exfoliated cells from cervix that could be present in urine protecting DNA. Moreover, the presence of DNA nucleases in plasma is much higher than in urine. In summary, this study suggests that only urine could serve as a possible replacement for cytobrush samples in CC detection, as long as panels of methylated genes are used together.
As cancer is a complex and dynamic disease that can change quickly, reliable and robust non-invasive platforms are needed for diagnosis in order to provide a personalized treatment. The liquid biopsy platforms described in the literature such as CTCs, exosomes, cfDNA, miRNAs, lncRNAs and other non-coding RNAs have the potential to add tremendous value to cancer patient care. The most important contribution of all forms of liquid biopsy lies in the detection of altered nucleic acids derived from tumors compared to the background of molecules derived from normal cells. Among these techniques, exosomes have a number of advantages for diagnostics, including obtaining a high-quality RNA from fresh or frozen biofluids, thus increasing the scope of detectable mutations, splice variants, fusions as well as expression-based assays for mRNA, miRNA, lncRNAs and other RNAs. Exosomes are also released from living cells as an active process, whereas cfDNA is released through the process of apoptosis and necrosis. Therefore, combining exosome RNA and cfDNA has the advantage of increasing the detection sensitivity for low frequency mutations (58).
For the patient there is an obvious and clear advantage to a liquid biopsy compared to conventional surgical methods. However, most of the current studies in biofluids have focused on the detection of actionable mutations more than on the methylation status of these deregulated genes. As DNA mutations will only provide information about some aspects of the disease, looking at RNA expression or methylation status of these genes in biofluids can help further understand the processes occurring in the body of a cancer patient.
The use of more modern technologies could increase the detection of DNA methylation in both blood and urine samples and thus improves the methodological sensitivity in detecting these epigenetic modifications. Some of these advances could involve techniques such as DROPLET Digital PCR, which is a more precise, sensitive and faster solution for a wide variety of applications, especially in the study of cancer biomarkers in different conditions due to its ability to measure several types of cancer mutations, detect rare DNA copies and epigenetic modifications, and detect markers with a low or variant number of copies in samples with superior sensitivity and resolution (59-61).
Conclusions
According to the results obtained, inconclusive evidence was found for using SFRP4 and ZAR1 hypermethylation for detecting SCC of CC in either urine or plasma. However, studies with larger numbers of participants and an improved sampling, mainly in urine, could increase the sensitivity of the detection of SFRP4 and ZAR1 methylation. Furthermore, the use of methylated gene panels would potentially allow early detection of CC and its precursor lesions in urine and plasma samples.
Acknowledgments
The authors wish to thank Claudia Diaz and Irene Riquelme for their invaluable help in recruiting patients and collecting samples for this study in the Gynecology outpatient service. The authors also thank Daniela León and Tamara Vizcarra for their help in laboratory procedures.
Funding: This work was supported by National Funding for Scientific and Technologic Development of Chile (FONDECYT) [3120141 to P.B., 11150802 to P.B., 11150622 to C.G.I]; Corporation for Production Development (CORFO) [No. 12IDL2-18157, No. 09CN14-5960 to CEGIN]; The Millennium Institute on Immunology and Immunotherapy No. P09-016-F; and CONICYT FONDAP No. 15130011.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.01.41). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All patients participated in the study signed an informed consent and the study approved by the School of Medicine Ethics Committee of Universidad de La Frontera (Approval No. 246/006).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bruni L, Barrionuevo-Rosas L, Serrano B, et al. ICO Information Centre on HPV and Cancer (HPV Information Centre). Human Papillomavirus and Related Diseases in World. Summary Report 2014-08-22: (2014). [Accessed 30th September 2014].
- Ferlay J, Soerjomataram I, Ervik M, et al. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [Internet]. Lyon, France: International Agency for Research on Cancer, 2013. Available online: http://globocan.iarc.fr, accessed on day/month/year
- Jung WW, Chun T, Sul D, et al. Strategies against human papillomavirus infection and cervical cancer. J Microbiol 2004;42:255-66. [PubMed]
- Burd EM. Human papillomavirus and cervical cancer. Clin Microbiol Rev 2003;16:1-17. [Crossref] [PubMed]
- Woodman CB, Collins SI, Young LS. The natural history of cervical HPV infection: unresolved issues. Nat Rev Cancer 2007;7:11-22. [Crossref] [PubMed]
- Muñoz N, Castellsagué X, de González AB, et al. Chapter 1: HPV in the etiology of human cancer. Vaccine 2006;24 Suppl 3:S3/1-10.
- Szalmás A, Kónya J. Epigenetic alterations in cervical carcinogenesis. Semin Cancer Biol 2009;19:144-52. [Crossref] [PubMed]
- Nguyen DX, Massagué J. Genetic determinants of cancer metastasis. Nat Rev Genet 2007;8:341-52. [Crossref] [PubMed]
- Momparler RL. Cancer epigenetics. Oncogene 2003;22:6479-83. [Crossref] [PubMed]
- Kitkumthorn N, Yanatatsanajit P, Kiatpongsan S, et al. Cyclin A1 promoter hypermethylation in human papillomavirus-associated cervical cancer. BMC Cancer 2006;6:55. [Crossref] [PubMed]
- Feng Q, Balasubramanian A, Hawes SE, et al. Detection of hypermethylated genes in women with and without cervical neoplasia. J Natl Cancer Inst 2005;97:273-82. [Crossref] [PubMed]
- Esteller M, Corn PG, Baylin SB, et al. A gene hypermethylation profile of human cancer. Cancer Res 2001;61:3225-9. [PubMed]
- Chung MT, Sytwu HK, Yan MD, et al. Promoter methylation of SFRPs gene family in cervical cancer. Gynecol Oncol 2009;112:301-6. [Crossref] [PubMed]
- Saavedra KP, Brebi PM, Roa JC. Epigenetic alterations in preneoplastic and neoplastic lesions of the cervix. Clin Epigenetics 2012;4:13. [Crossref] [PubMed]
- Hoque MO, Topaloglu O, Begum S, et al. Quantitative methylation-specific polymerase chain reaction gene patterns in urine sediment distinguish prostate cancer patients from control subjects. J Clin Oncol 2005;23:6569-75. [Crossref] [PubMed]
- Goessl C, Müller M, Straub B, et al. DNA alterations in body fluids as molecular tumor markers for urological malignancies. Eur Urol 2002;41:668-76. [Crossref] [PubMed]
- Shivapurkar N, Gazdar AF. DNA methylation based biomarkers in non-invasive cancer screening. Curr Mol Med 2010;10:123-32. [Crossref] [PubMed]
- Yu J, Zhu T, Wang Z, et al. A novel set of DNA methylation markers in urine sediments for sensitive/specific detection of bladder cancer. Clin Cancer Res 2007;13:7296-304. [Crossref] [PubMed]
- Chung W, Bondaruk J, Jelinek J, et al. Detection of bladder cancer using novel DNA methylation biomarkers in urine sediments. Cancer Epidemiol Biomarkers Prev 2011;20:1483-91. [Crossref] [PubMed]
- Goessl C, Müller M, Heicappell R, et al. DNA-based detection of prostate cancer in urine after prostatic massage. Urology 2001;58:335-8. [Crossref] [PubMed]
- Rogers CG, Gonzalgo ML, Yan G, et al. High concordance of gene methylation in post-digital rectal examination and post-biopsy urine samples for prostate cancer detection. J Urol 2006;176:2280-4. [Crossref] [PubMed]
- Feng Q, Hawes SE, Stern JE, et al. Promoter hypermethylation of tumor suppressor genes in urine from patients with cervical neoplasia. Cancer Epidemiol Biomarkers Prev 2007;16:1178-84. [Crossref] [PubMed]
- Begum S, Brait M, Dasgupta S, et al. An epigenetic marker panel for detection of lung cancer using cell-free serum DNA. Clin Cancer Res 2011;17:4494-503. [Crossref] [PubMed]
- Lange CP, Campan M, Hinoue T, et al. Genome-scale discovery of DNA-methylation biomarkers for blood-based detection of colorectal cancer. PLoS One 2012;7:e50266 [Crossref] [PubMed]
- Müller HM, Fiegl H, Widschwendter A, et al. Prognostic DNA methylation marker in serum of cancer patients. Ann N Y Acad Sci 2004;1022:44-9. [Crossref] [PubMed]
- Widschwendter A, Müller HM, Fiegl H, et al. DNA methylation in serum and tumors of cervical cancer patients. Clin Cancer Res 2004;10:565-71. [Crossref] [PubMed]
- Yang HJ, Liu VW, Wang Y, et al. Detection of hypermethylated genes in tumor and plasma of cervical cancer patients. Gynecol Oncol 2004;93:435-40. [Crossref] [PubMed]
- Ren CC, Miao XH, Yang B, et al. Methylation status of the fragile histidine triad and E-cadherin genes in plasma of cervical cancer patients. Int J Gynecol Cancer 2006;16:1862-7. [Crossref] [PubMed]
- Abudukadeer A, Bakry R, Goebel G, et al. Clinical Relevance of CDH1 and CDH13 DNA-Methylation in Serum of Cervical Cancer Patients. Int J Mol Sci 2012;13:8353-63. [Crossref] [PubMed]
- Brebi P, Maldonado L, Noordhuis MG, et al. Genome-wide methylation profiling reveals Zinc finger protein 516 (ZNF516) and FK-506-binding protein 6 (FKBP6) promoters frequently methylated in cervical neoplasia, associated with HPV status and ethnicity in a Chilean population. Epigenetics 2014;9:308-17. [Crossref] [PubMed]
- Brebi P, Hoffstetter R, Andana A, et al. Evaluation of ZAR1 and SFRP4 methylation status as potentials biomarkers for diagnosis in cervical cancer: exploratory study phase I. Biomarkers 2014;19:181-8. [Crossref] [PubMed]
- Aedo AS, Melo AA, García P, et al. Detection and typification of human papilloma virus in pre cancerous cervical lesions. Rev Med Chil 2007;135:167-73. [PubMed]
- Swets JA. Measuring the accuracy of diagnostic systems. Science 1988;240:1285-93. [Crossref] [PubMed]
- Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010;127:2893-917. [Crossref] [PubMed]
- Schatz P, Dietrich D, Koenig T, et al. Development of a diagnostic microarray assay to assess the risk of recurrence of prostate cancer based on PITX2 DNA methylation. J Mol Diagn 2010;12:345-53. [Crossref] [PubMed]
- Semczuk A, Jakowicki JA. Alterations of pRb1-cyclin D1-cdk4/6-p16INK4A pathway in endometrial carcinogenesis. Cancer Lett 2004;203:1-12. [Crossref] [PubMed]
- Dong SM, Kim HS, Rha SH, et al. Promoter Hypermethylation of Multiple Genes in Carcinoma of the Uterine Cervix. Clin Cancer Res 2001;7:1982-6. [PubMed]
- Sellors JW, Lorincz AT, Mahony JB, et al. Comparison of self-collected vaginal, vulvar and urine samples with physician-collected cervical samples for human papillomavirus testing to detect high-grade squamous intraepithelial lesions. CMAJ 2000;163:513-8. [PubMed]
- Vorsters A, Micalessi I, Bilcke J, et al. Detection of human papillomavirus DNA in urine. A review of the literature. Eur J Clin Microbiol Infect Dis 2012;31:627-40. [Crossref] [PubMed]
- Enerly E, Olofsson C, Nygård M. Monitoring human papillomavirus prevalence in urine samples: a review. Clin Epidemiol 2013;5:67-79. [Crossref] [PubMed]
- Rouprêt M, Hupertan V, Yates DR, et al. Molecular Detection of Localized Prostate Cancer Using Quantitative Methylation-Specific PCR on Urinary Cells Obtained Following Prostate Massage. Clin Cancer Res 2007;13:1720-5. [Crossref] [PubMed]
- Su YH, Wang M, Block TM, et al. Transrenal DNA as a diagnostic tool: important technical notes. Ann N Y Acad Sci 2004;1022:81-9. [Crossref] [PubMed]
- Umansky SR, Tomei LD. Transrenal DNA testing: progress and perspectives. Expert Rev Mol Diagn 2006;6:153-63. [Crossref] [PubMed]
- Petrucci R, Lombardi G, Corsini I, et al. Use of transrenal DNA for the diagnosis of extrapulmonary tuberculosis in children: a case of tubercular otitis media. J Clin Microbiol 2015;53:336-8. [Crossref] [PubMed]
- Khan G, Kangro HO, Coates PJ, et al. Inhibitory effects of urine on the polymerase chain reaction for cytomegalovirus DNA. J Clin Pathol 1991;44:360-5. [Crossref] [PubMed]
- Johnson DJ, Calderaro AC, Roberts KA. Variation in nuclear DNA concentrations during urination. J Forensic Sci 2007;52:110-3. [Crossref] [PubMed]
- Hoque MO, Feng Q, Toure P, et al. Detection of aberrant methylation of four genes in plasma DNA for the detection of breast cancer. J Clin Oncol 2006;24:4262-9. [Crossref] [PubMed]
- Brooks JD, Cairns P, Shore RE, et al. DNA methylation in pre-diagnostic serum samples of breast cancer cases: results of a nested case-control study. Cancer Epidemiol 2010;34:717-23. [Crossref] [PubMed]
- Herman JG. Circulating methylated DNA. Ann N Y Acad Sci 2004;1022:33-9. [Crossref] [PubMed]
- Patel A, Groopman JD, Umar A. DNA methylation as a cancer-specific biomarker: from molecules to populations. Ann N Y Acad Sci 2003;983:286-97. [Crossref] [PubMed]
- Li L, Choi JY, Lee KM, et al. DNA methylation in peripheral blood: a potential biomarker for cancer molecular epidemiology. J Epidemiol 2012;22:384-94. [Crossref] [PubMed]
- Kindermann G, Jabusch HP. The spread of squamous cell carcinoma of the uterine cervix into the blood-vessels. Arch Gynakol 1972;212:1-8. [Crossref] [PubMed]
- Fleischhacker M, Schmidt B. Circulating nucleic acids (CNAs) and cancer--a survey. Biochim Biophys Acta 2007;1775:181-232.
- McKay JA, Xie L, Harris S, et al. Blood as a surrogate marker for tissue-specific DNA methylation and changes due to folate depletion in post-partum female mice. Mol Nutr Food Res 2011;55:1026-35. [Crossref] [PubMed]
- Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 2013;200:373-83. [Crossref] [PubMed]
- Zhang X, Yuan X, Shi H, et al. Exosomes in cancer: small particle, big player. J Hematol Oncol 2015;8:83. [Crossref] [PubMed]
- Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics 1977;33:159-74. [Crossref] [PubMed]
- Brock G, Castellanos-Rizaldos E, Hu L, et al. Liquid biopsy for cancer screening, patient stratification and monitoring. Transl Cancer Res 2015;4:280-90.
- Hindson BJ, Ness KD, Masquelier DA, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 2011;83:8604-10. [Crossref] [PubMed]
- Hindson CM, Chevillet JR, Briggs HA, et al. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods 2013;10:1003-5. [Crossref] [PubMed]
- Jones M, Williams J, Gärtner K, et al. Low copy target detection by Droplet Digital PCR through application of a novel open access bioinformatic pipeline, “definetherain J Virol Methods 2014;202:46-53. [Crossref] [PubMed]