
Identification the potential of stool-based SNCA methylation as a candidate biomarker for early colorectal cancer detection
Introduction
Colorectal cancer (CRC) has become one of the leading causes of cancer-associated death worldwide. According to the pathological study, CRC mainly arises from benign precursors (adenomas) to cancer during a long time interval. Detection of CRC at an early curable stage and removal of precancerous lesions can improve the cure rate (1). At present, the conventional methods, fecal occult blood test (FOBT) and colonoscopy are used most frequently to screen for CRC. However, FOBT might be interfered by diet composition and the measurement repeatability is limited. Colonoscopy is invasive, costly and low adherence rates, although it has highly positive predictive value (1). Therefore, it is imperative to develop molecular biomarkers as a noninvasive screening method for the diagnosis of early CRC.
Hypermethylation of DNA promoter occurs early in the development of CRC (2), suggesting its suitability as a molecular marker in early diagnosis of CRC. Periodic exfoliation of intestinal epithelial cells provides a basis for the analysis of stool DNA to identify methylation variation of intestinal tissue (3-6). But previous studies on stool DNA methylation showed lower sensitivity for detecting adenomas or advanced precancerous lesions which would be the curable stage to avoid the morbidity of CRC (3,5). In addition, another crucial factor influencing test outcomes is stool sample processing to recover analyzable human DNA from stool (7).
Alpha-synuclein (SNCA) gene encodes conserved protein that is abundant in neurons, especially presynaptic terminals. Aggregated alpha-synuclein proteins form brain lesions that are hallmarks of neurodegenerative synucleinopathies (8). Previous studies have shown SNCA hypermethylation in blood and cerebral cortex of patients with Parkinsons’s disease (9), in biliary brush samples with cholangiocarcinoma (10), and in CRC cell lines (11). Spastic paraplegia-20 (SPG20) gene encodes the multifunctional spartin protein, which was found to be involved in intracellular epidermal growth factor receptor trafficking (12). SPG20 was demonstrated to be methylated in 89%, 78% and 1% of the CRC, adenomas and normal mucosa samples, respectively (13). Fibrillin-1 (FBN1) encodes the structural fibrillin protein, which is an extracellular matrix glycoprotein that serves as a structural component of calcium-binding microfibrils. FBN1 was hypermethylated in CRC samples compared to the normal colorectal mucosa (11). Our previous work in tissue samples showed hypermethylation of the promoter of SNCA, SPG20, and FBN1 in CRC patients (14).
In this study, we aimed to explore the potential of stool-based DNA methylation of the three genes (SNCA, SPG20, FBN1) as suitable candidate biomarkers to discriminate patients with adenomas and colorectal cancer from normal controls and to illuminate the relationship between validated biomarkers and clinicopathologic factors.
Methods
Clinical samples
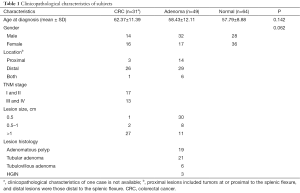
Thirty-one patients with CRC, 49 patients with adenomas, and 64 normal controls were recruited in the National Center of Colorectal Surgery, the 3rd affiliated Hospital of Nanjing University of Traditional Chinese Medicine between April 2014 and February 2015. Details regarding the following information are summarized in Table 1: gender, age of onset, tumor location, lesion size, histological and TNM staging classifications. There were no age and gender differences between CRC/adenomas groups and normal controls. The adenomas included adenomatous polyp, tubular adenomas (TA), tubulovillous adenomas (TVA), and high-grade intraepithelial neoplasm (HGIN). An advanced precancerous lesion was defined as tubular adenomas 1cm or larger, TVA, and HGIN (15). All participants in the study provided written informed consent and this study were approved by the Ethics Committee of the 3rd affiliated Hospital of Nanjing University of Traditional Chinese Medicine (KY2014018).
Full table
Stool processing using the stool intestinal mucosal cells collector
All control/adenomas stool samples and a subset of the CRC stool samples were collected before bowel purgation and colonoscopy. Some CRC stool samples were collected >7 days after colonoscopy. A single stool sample was put into the provided collection container and homogenized in a DNA stabilization buffer and frozen at −80 °C immediately. The stored stool samples were further processed within 3 days. All stool samples were handled in a blinded fashion during storage, processing, DNA extraction, and qMSP analysis.
To obtain high quality and quantity of intestinal mucosal cells from stool samples, a stool device called the stool intestinal mucosal cells collector was designed. This device consists of three volumes of 1,000 mL containers containing the filter screen on the bottom with pore diameter of 100, 200, and 300 meshes, respectively (Figure 1).
The operational steps of the stool processing were as follows: Stool sample (20–30 g) mixed with 500 mL of buffer was poured into the container A. Vibrate the container to let the mixture passes through filter screens of container A, B, and C in sequence. The filter liquor were collected and poured into a 500 mL centrifuge tube and centrifuge at 5,000 rpm for 20 min. Discard the supernatant and transfer the middle layer (intestinal mucosal cells enrichment) into a new 50 mL centrifuge tube and store at −80 °C.
DNA extraction and bisulfite modification
Genomic DNA in stool extractions was obtained using the TIANamp Genomic DNA kit (Qiagen) according to the manufacturer’s instructions. DNA concentration was around 100 µg/mL and purity (OD260/280) was around 1.8, suggesting the stool DNA was achieved for the requirement of DNA methylation detection.
DNA (1–2 µg) was bisulfite modified using CpGenomeTM Universal DNA Modification Kit (Millipore) according to manufacturer’s protocol and eluted in 20 µL of TE buffer (stool sample). CpGenomeTM Universal Methylated DNA (Millipore) was used as a methylation-positive control.
Quantitative methylation-specific polymerase chain reaction (qMSP)
The methylation status of SNCA, SPG20, and FBN1 in stool samples was confirmed by qMSP. The primer and probe sequences are listed in Table S1. The qMSP was carried out in a 10 µL reaction volume including 0.7 µL each of forward and reverse primers (0.9 µM), 0.4 µL probe (0.4 µM), 3 µL bisulfite treated template, 5 µL TaKaRa Premix Ex TaqTM and 0.2 µL ROX Reference Dye. The PCR program was performed by ABI StepOne Plus real-time PCR system (Applied Biosystems) as follows: 30 sec at 95 °C, then 55 cycle of 10 sec at 95 °C and 30 sec at 55 °C. The qMSP results were calculated as percentage of methylated reference [PMR, the GENE: ALU ratio of a sample was divided by the GENE: ALU ratio of the positive control (CpGenome Universally Methylated DNA) and multiplied by 100] (11). The most optimal PMR value was determined at the point on the ROC curve at which (sensitivity + specificity) was maximal.
Statistical analysis
This study was planned to get a power of 80% to test the hypothesis that the DNA methylation test would achieve a sensitivity of 75% or more for the detection of CRC under the null hypothesis, at a one sided type I error rate of 0.05. PMR Differences in stool SNCA methylation status between two independent groups were analyzed by Mann-Whitney U test, and between paired samples by Wilcoxon signed-rank. PMR Differences among groups more than two were analyzed by Kruskal-Wallis H test. Detection rates differences were compared by Fisher’s exact test. ROC curve was generated using PMR values. For the ROC curve analysis, the areas under the curve (AUC) values were used to reflect the diagnostic performance of SNCA methylation status. The P values for the ROC curves are generated under the nonparametric assumption with the null hypothesis: true area =0.5. Multivariable logistic analyses were used to calculate the odds ratio (OR) related to CRC or adenomas according to stool SNCA methylation levels after adjustment of age and sex cases. The statistical tests were performed by the SPSS software package (version 16). P<0.05 was considered statistically significant.
Results
Significant difference in SNCA methylation between CRC/adenomas groups and normal controls in stool sample
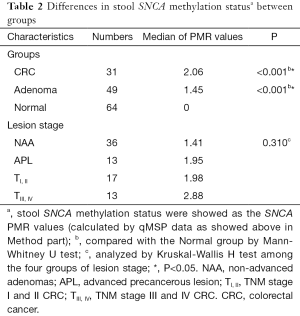
SNCA, SPG20 and FBN1 promoter methylation were evaluated by qMSP in 144 stool samples including 31 CRCs, 49 adenomas, and 64 normal controls. The SNCA PMR values (calculated by qMSP data as showed above in Method part) was significantly higher in CRC (median 2.06) and adenomas (median 1.45) than the normal control (median 0) (P<0.001 and P<0.001, respectively. Table 2 and Figure 2A). However, there were no significant differences in stool SPG20 or FBN1 methylation among CRC, adenomas and normal controls (P>0.05).
Full table

ROC curves were used to evaluate the discriminating potential of SNCA methylation as a noninvasive biomarker for detecting CRC and adenomas. The area under the ROC curve (AUC) values for SNCA were 0.836 (P<0.001) and 0.772 (P<0.001) for the detection of CRC and adenomas, respectively (Figure 2B,C). The optimal cutoff value for scoring positive methylation was set at PMR of 0.74 using method showed above in Method part. The sensitivity was 83.9% and 75.5% to identify a patient with CRC and adenomas, respectively and the specificity was 75% for both (Table 3).

Full table
In addition, multivariable logistic regression analyses showed that the OR for CRC and adenomas patients with high stool-based SNCA methylation was 11.291 and 9.234, respectively, after adjustment for patients’ age and sex (Table 4). Thus, it revealed that SNCA methylation could be used as a potential biomarker for identifying patients with CRC and adenomas. Table 4 also showed that the elder age can increase the prevalence risk of CRC.

Full table
SNCA methylation showed an increasing trend across the disease transition but the difference was not significant
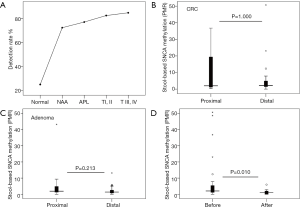
PMR values of stool-based SNCA methylation showed an increasing trend across the disease transition from non-advanced adenomas (NAA, n=36), advanced precancerous lesion (APL, n=13), tumor-node-metastasis (TNM) stage I and II CRC (n=17) to TNM stage I and II CRC (n=13), but the difference was not significant (P=0.310, Table 2). When compared PMR of CRC TNM stage I and II with TNM stage I and II, or NAA with APL, the differences were not significant as well (P=0.660 and 0.248, respectively). Detection rates of stool-based SNCA methylation according to PMR cut-off showed a stepwise increasing trend across the disease transition (72.2% for NAA, 76.9% for APL, 82.3% for TNM stage I and II CRC, and 84.6% for TNM stage I and II CRC, Figure 3A), but the difference was not significant (P=0.788 by Fisher’s exact test).
Stool-based SNCA methylation was not associated with the location of CRC or adenomas
To determine the detection rates of CRC or adenomas according to the location within the colorectum, SNCA methylation between proximal and distal lesions was compared. The results illustrated that stool-based SNCA methylation did not correlate with lesion location from patients with CRC (P=1.000, Figure 3B) or adenomas (P=0.213, Figure 3C).
Stool-based SNCA methylation is reduced after tumor resection
Thereafter, we intended to determine whether stool-based SNCA methylation was altered subsequent to tumor resection in stool CRC sample (31 before vs. after). It’s interesting to note that SNCA methylation significantly decreased after tumor resection compared with the initial status (P=0.010, Figure 3D).
Discussion
In this study, we explored the potential of stool-based DNA methylation as biomarkers for discriminating CRC or adenomas from normal controls.
The challenge of detecting stool DNA is to recover trace amounts of analyzable human DNA. In stool, only a tiny proportion of exfoliation of intestinal mucosal cells are mixed with a large amount of bacteria and diet, which increase the difficulty to get enrichment of stool human DNA. Our data indicated the high quality of stool DNA processed by the stool intestinal mucosal cells collector, suggesting the obtained stool DNA was satisfied for the requirement of DNA methylation detection.
Evidence from our previous and other studies indicated methylation of SNCA in >80% of CRC cell lines and the frequency of SNCA methylation was significantly higher in CRC tissues compared to adjacent normal ones (11,14). To our knowledge, this study is the first to investigate the potential of SNCA methylation in stool samples as biomarkers. The results showed that stool-based SNCA methylation had highly discriminatory performance for detecting CRC or adenomas. This is supported by AUC values of 0.836 for the detection of CRC (sensitivity =83.9%; specificity =75%) and 0.772 for adenomas (sensitivity =75.5%; specificity =75%). Moreover, the OR for CRC and adenomas with stool SNCA hypermethylation was 11.291 and 9.234, respectively. In addition, logistic analyses showed a high prevalence of risk factors for CRC in persons 65 years or older compared to the ones less than 65 years old.
As we know, to reduce the mortality of CRC, diagnosis of the disease at curable stages, such as adenomas, precancerous lesions and early CRC, is highly important (16). Yet, the previous reports mostly refer to CRC state, and the detecting sensitivity is lower for adenomas (3,5). Our current study showed stool-based SNCA methylation had a sensitivity of 80% for early colorectal neoplasm (including advanced precancerous lesion and CRC at stage I and II). This may provide an important opportunity for the ideal stage of early detection of CRC. Another finding of our study was the significant decrease in stool-based SNCA methylation levels after tumor resection in patients with CRC, which indicated that SNCA methylation is a specific biomarker for CRC. Though a trend of growing PMR of stool-based SNCA methylation found through the adenoma-cancer sequence (Figure 3A), the difference was not significant. This is perhaps because of the relative small sample size. Or molecular alterations of CRC might occur earlier since the initial of adenoma.
Stool SPG20 and FBN1 methylation status did not differ significantly among CRC, adenomas and normal controls in our study, suggesting the two genes are not suitable for CRC diagnosis.
Our present findings have several clinical implications. First, though colonoscopy is a gold standard for detecting CRC, it’s invasive. In addition, molecular alterations of CRC might occur earlier than histomorphology changes, which suggest that a test using stool-based DNA methylation may provide noninvasive detection at lesions prior to malignancy. Second, sensitivity is an important feature for screening tests. In our study, the sensitivity (80%) of stool-based SNCA methylation for the detection of early colorectal neoplasm exceeded the performance of many previous reports. Third, as the relative incidence of proximal CRC and adenomas is higher than those of distal, effective detection of proximal lesions is of great importance. Previous studies demonstrated that colonoscopy is less effective for proximal than distal colon neoplasms (17,18), while stool-based SNCA methylation has comparable efficacy in the proximal and distal colorectum for detection of both CRC and adenomas. In view of this performance, stool-based SNCA methylation testing has potential to extend the scope for detecting neoplasms throughout the colorectum.
The limitations of this study include the following. First, patients were from a single center and some were symptomatic, and larger scale validation across multiple centers and different populations is required. Second, the specificity of the stool-based SNCA methylation (75%) is inferior to that of the multitarget stool DNA test (86.6–89%) (6). A combined biomarker panel may increase the specificity in screening compared to a single marker (11). Third, the sample size is relative small, especially for the proximal CRC group, and this will reduce the test power of this study.
Conclusions
Our results highlight the capability of stool-based SNCA methylation to detect adenomas which provide rationale for further development of the epigenetic biomarker in noninvasive early detection of CRC. Our next step is to detect combing molecular assays with better diagnostic performance for the early detection of CRC.

Full table
Acknowledgments
Funding: This study was supported by National Nature Science Foundation of China (No. 30572447, 30973837, 81273944) and Natural Science Foundation of Jiangsu, China (BK2012724).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.01.30). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All participants in the study provided written informed consent and this study were approved by the Ethics Committee of the 3rd affiliated Hospital of Nanjing University of Traditional Chinese Medicine (KY2014018).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Levin B, Lieberman DA, McFarland B, et al. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. CA Cancer J Clin 2008;58:130-60. [Crossref] [PubMed]
- Shames DS, Minna JD, Gazdar AF. DNA methylation in health, disease, and cancer. Curr Mol Med 2007;7:85-102. [Crossref] [PubMed]
- Glöckner SC, Dhir M, Yi JM, et al. Methylation of TFPI2 in stool DNA: a potential novel biomarker for the detection of colorectal cancer. Cancer Res 2009;69:4691-9. [Crossref] [PubMed]
- Bosch LJ, Oort FA, Neerincx M, et al. DNA methylation of phosphatase and actin regulator 3 detects colorectal cancer in stool and complements FIT. Cancer Prev Res (Phila) 2012;5:464-72. [Crossref] [PubMed]
- Ahlquist DA, Zou H, Domanico M, et al. Next-generation stool DNA test accurately detects colorectal cancer and large adenomas. Gastroenterology 2012;142:248-56; quiz e25-6.
- Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med 2014;370:1287-97. [Crossref] [PubMed]
- Osborn NK, Ahlquist DA. Stool screening for colorectal cancer: molecular approaches. Gastroenterology 2005;128:192-206. [Crossref] [PubMed]
- Giasson BI, Duda JE, Murray IV, et al. Oxidative damage linked to neurodegeneration by selective alpha-synuclein nitration in synucleinopathy lesions. Science 2000;290:985-9. [Crossref] [PubMed]
- Pihlstrøm L, Berge V, Rengmark A, et al. Parkinson's disease correlates with promoter methylation in the α-synuclein gene. Mov Disord 2015;30:577-80. [Crossref] [PubMed]
- Andresen K, Boberg KM, Vedeld HM, et al. Four DNA methylation biomarkers in biliary brush samples accurately identify the presence of cholangiocarcinoma. Hepatology 2015;61:1651-9. [Crossref] [PubMed]
- Lind GE, Danielsen SA, Ahlquist T, et al. Identification of an epigenetic biomarker panel with high sensitivity and specificity for colorectal cancer and adenomas. Mol Cancer 2011;10:85. [Crossref] [PubMed]
- Bakowska JC, Jupille H, Fatheddin P, et al. Troyer syndrome protein spartin is mono-ubiquitinated and functions in EGF receptor trafficking. Mol Biol Cell 2007;18:1683-92. [Crossref] [PubMed]
- Lind GE, Raiborg C, Danielsen SA, et al. SPG20, a novel biomarker for early detection of colorectal cancer, encodes a regulator of cytokinesis. Oncogene 2011;30:3967-78. [Crossref] [PubMed]
- Ma J, Yang Q, Deng C, et al. Screening and diagnostic value of the molecular markers of DNA methylation in colorectal neoplasma. Zhonghua Wei Chang Wai Ke Za Zhi 2015;18:1149-53. [PubMed]
- Yamada A, Horimatsu T, Okugawa Y, et al. Serum miR-21, miR-29a, and miR-125b Are Promising Biomarkers for the Early Detection of Colorectal Neoplasia. Clin Cancer Res 2015;21:4234-42. [Crossref] [PubMed]
- Kim DH, Pickhardt PJ, Taylor AJ, et al. CT colonography versus colonoscopy for the detection of advanced neoplasia. N Engl J Med 2007;357:1403-12. [Crossref] [PubMed]
- Brenner H, Hoffmeister M, Arndt V, et al. Protection from right- and left-sided colorectal neoplasms after colonoscopy: population-based study. J Natl Cancer Inst 2010;102:89-95. [Crossref] [PubMed]
- Baxter NN, Goldwasser MA, Paszat LF, et al. Association of colonoscopy and death from colorectal cancer. Ann Intern Med 2009;150:1-8. [Crossref] [PubMed]



