
DNA methylation biomarkers in lung cancer diagnosis: closer to practical use?
The purpose of cancer screening is to identify those patients for whom survival might be improved by early intervention. The strategy of waiting for symptoms results in a greater proportion of patients with advanced disease, fewer treated with curative intent and even fewer surviving long term. In some patients, such as the very elderly or infirmed, lung cancer may not shorten survival, and screening in them is less likely beneficial.
The National Lung Screening Trial (NLST) performed three annual computed tomogram (CT) screens and had established trial guidelines for managing discovered lesions (1). By design, all the participating institutions had high quality systems to manage discovered abnormalities. The low risk results are unlikely to be matched in the real world; the psychological toll, cost, morbidity, and mortality are more likely to be a burden on society. So, developing an additional test, such as from the blood or sputum, would be of benefit. Many have attempted to improve the diagnostic capabilities of imaging using blood, plasma, serum, sputum, buccal smears, and breath analysis. The best results have had marginal improvement and were impractical to perform.
Combining additional clinical information may improve the sensitivity and specificity of a testing system. For example, important parameters may include age, smoking pack-years, COPD status, forced vital capacity, lesion diameter, density, location in the upper lobe versus other lobes, border characteristics, presence of corona radiata, pleural retraction, presence and character of calcifications, possible contrast enhancement, presence of emphysema, emphysematous changes, to name a few. It remains unclear how this data provides additional help in ruling in our ruling out cancer.
The next concern is typical of screening studies: who are the patients at risk? Yes, smokers with greater than 15 pack-years and older patients are at risk for lung cancer, but there are other patients that may have significant risk factors other than smoking. Those not included in the NLST and other similar trials include patients with inflammatory lung disease, generalized inflammatory diseases such as rheumatoid arthritis, genetic predisposition, and exposures to toxins, such as asbestos. By choosing a test group, the authors may not be studying the patients most in need of an additional screening test.
DNA methylation abnormalities are found in most human tumors including lung cancer. DNA methyltransferase enzymes methylate between 70% and 80% of the CpG dinucleotide sequences in mammalian DNA at position 5 of cytosine. Cytosine methylation is an epigenetic modification that is generally incompatible with gene expression when present at gene promoters. However, there are genomic landmarks that are particularly rich in CpG sequences, yet they are kept in an unmethylated state during organismal development, in most somatic tissues and in the germ line (2). Specialized proteins and mechanisms may keep these CpG islands free of DNA methylation (3) but their modus of operation have not yet been well defined. These protective barriers break down during the development of cancer leading to the commonly observed phenomenon of CpG island hypermethylation in tumors (4). DNA hypermethylation is observed in every malignancy tested, and is found even at early stages of tumor progression. In lung cancer, several hundred and up to a thousand CpG islands undergo methylation relative to normal lung tissue of the same patient (5-8).
Although numerous methylation changes have been catalogued, it has been difficult to discern which ones of these DNA hypermethylation events in cancer have the properties of being tumor-driving events. This challenge is not unlike the one we face with tumor-associated mutations of which we know that hundreds or thousands of mutations exist in an individual tumor genome but based on our current knowledge we can only call out a handful of them as validated tumor drivers. Defining DNA methylation changes as tumor driving is complicated by the fact that they likely occur through a methylation targeting mechanism rather than by selection of a tumor-promoting phenotype. Very often, a particularly class of genes referred to as Polycomb targets undergoes widespread methylation (6,9-12), not only in lung cancer but also in many other tumors. These genes are occupied and modified by the Polycomb repression complex in normal cells including stem cells, which—through an unknown mechanism—creates a strong susceptibility for DNA methylation to occur in tumors. Perhaps it is the large number of DNA hypermethylation events occurring simultaneously that provides a growth advantage to the cell. For small cell lung cancer we observed that DNA methylation leads to a potential defect in cell differentiation that promotes malignant transformation (5). There are numerous studies that have reported a worse clinical outcome when DNA hypermethylation occurs, both at the level of individual genes or for groups of genes combined (13-15).
Regardless of the biological meaning of CpG island hypermethylation in cancer, the DNA methylation events in cancer are specific for the malignant state and do not occur, or occur at much lower frequency in normal tumor-adjacent tissue or in the normal cell type from which the cancer is thought to originate. Based on these observations, DNA methylation changes in cancer have long been considered as powerful potential biomarkers of the disease (16-20). These methylation biomarkers could be useful for early detection, classification of cancer subtypes, clinical outcome predictions, or even disease management and treatment choices. Just to give one example for tumor classification, the presence of the “CpG island methylator phenotype” (CIMP), which is based on the presence of an unusually large number of CpG island methylation events in a subset of patients (21), is a common occurrence in subgroups of colorectal cancers and brain cancers. In some instances, CIMP has been associated with specific genetic changes in the same tumor specimens. For CIMP in colorectal tumors, the presence of a BRAF mutation has been noted (22), although the exact mechanism how these two events within the genetic and epigenetic landscapes of tumors are linked has remained unclear. IDH1 mutations in lower grade gliomas are also associated with CIMP (23). In this case, the mutant IDH1 protein (commonly IDH1 R132H) produces high levels of the metabolite 2-hydroxyglutarate which is a competitive inhibitor of 2-ketoglutarate-dependent dioxygenase enzymes. One popular theory is that 2-hydroxyglutarate inhibits 5-methylcytosine oxidases, the TET enzymes, thus leading to an imbalance of DNA methylation patterns in IDH1 mutant brain tumors.
However, even in the absence of CIMP, most tumors carry several hundred to a few thousand CpG island hypermethylation events within individual tumor specimens. Thus it is generally not too difficult to screen for, identify and then characterize CpG islands that can serve as potential DNA methylation markers. Such a marker needs to fulfill a number of criteria: (I) it should be very commonly methylated in a series of patients with the same malignancy (ideally in all patients); (II) the methylation level, i.e., the frequency of methylated alleles at the target locus, should be as high as possible in the tumor; (III) background methylation levels in normal tissues, both in the target organ and in other healthy tissues that might be present in the analytical specimens should be close to zero. For example, if buccal cells were to be used for methylation analysis, methylation at the biomarker CpG island should be close to zero in normal buccal epithelial cells but also in lymphocytes and other immune cells that may be present in the buccal mucosa.
Having identified a methylation biomarker that fulfills these criteria, the next challenge is to develop a highly sensitive assay for its detection. One advantage of a DNA hypermethylation marker is that it provides a positive signal that might stand out from a large background of having no signal. Ideally the diagnostic test would be a noninvasive one that does not require tissue biopsy or other unpleasant or risky procedure such as bronchoscopy. Although the latter procedures are likely to sample a substantial fraction of tumor cells for lung cancer diagnosis, they may be difficult to implement in the clinical practice for screening a larger population. Therefore, testing for methylation biomarkers in serum, plasma or sputum have been considered as viable alternatives for lung cancer screening. The main challenge with this approach is the low amounts of tumor-derived DNA found in body fluids such as serum (24). The level of serum DNA is often increased in tumor patients relative to normal healthy individuals. However, the amounts and the integrity of DNA fragments seem to vary considerably between patients. This could depend on a number of parameters, most notably on whether the tumor is present in an early or late stage of malignant progression. Tumor-derived DNA may also be present in sputum and in this case it is likely that tumors near the central airway system would shed more tumor cells into sputum than more peripherally located tumors. With sputum, it may also be difficult to separate tumor cells from normal epithelial cells. Furthermore, sputum samples are much more readily obtained from current smokers than from nonsmokers or from those who have quit smoking a long time ago.
The first studies on lung cancer diagnosis using DNA methylation markers were reported almost two decades ago (25,26). The number of patients was small but in one study it was possible to detect methylated DNA sequences in sputum 3 years prior to clinical diagnosis of a lung tumor (27). Lack of sufficient specificity and problems with the sensitivity of the assays has made it difficult to advance these studies into the clinical practice.
A new report by Hulbert et al. published in Clinical Cancer Research now describes important progress in diagnosing lung cancer using DNA methylation markers (28). The authors designed a case-control study of individuals with suspicious nodules detected by CT imaging. Plasma and sputum were analyzed before surgery. The study included 150 cases of non-small cell lung cancer confirmed by pathology. They were all node negative (stage I and II). The 60 controls had no cancer diagnosis upon pathological examination.
Hulbert et al. used a technique with increased sensitivity “methylation on beads” that was designed to minimize sample loss. A method with extremely high sensitivity is paramount for implementing methylation diagnostics. One of the most sensitive methods is methylation-specific PCR, which uses PCR primers that distinguish between methylated and unmethylated alleles after bisulfite conversion (29). This conversion deaminates cytosine to uracil, which later amplifies as T, but bisulfite cannot deaminate 5-methylcytosine, which amplifies as C. Used in the format of quantitative real-time PCR, this method is very sensitive and specific. To ensure a maximum level of specific amplification of the locus in tumor DNA but not in normal cells or tissue, the methylation state at the queried CpG sites should show the greatest differences possible (see Figure 1). The opposing methylation state in tumor versus normal tissue DNA could be confirmed by high throughput bisulfite sequencing of the targeted region.
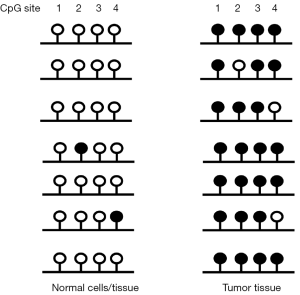
The authors started from publically available information to identify six DNA methylation markers that are methylated in a large fraction of patient cohorts, for example as published by The Cancer Genome Atlas (TCGA). These six genes included SOX17, TAC1, HOXA7, HOXA9, CDO1, and ZFP42. The gene set included the homeobox genes HOXA7 and HOXA9, which are very frequently methylated in non-small cell lung cancers as previously reported (7). There is a large set of useful DNA methylation markers to choose from inasmuch as other groups have used different sets of very specific markers to detect lung cancer in sputum (30). Using criteria for highest sensitivity and specificity in patients versus controls, Hulbert et al. narrowed the gene list down to three, TAC1, HOXA7 and SOX17. With sputum testing, the positive and negative predictive values for these genes were 93% and 58% for TAC1, 97% and 40% for HOXA7 and 96% and 60% for SOX17, respectively (28). For plasma, the gene CDO1 provided better data than TAC1 and was therefore used. The positive and negative predictive values were 86% and 46% for CDO1, 90% and 57% for TAC1, and 92% and 55% for SOX17. Based on these two gene sets, it appears that overall comparable predictions were possible with sputum and plasma as testing material. When the authors included only smokers in their analysis, similar results were obtained. Employment of blinded random forest prediction models also showed that methylation values were more important variables than demographic and clinical variables alone. Using sputum analysis, for example, the authors found that the random forest model with methylation markers correctly predicted lung cancer in 91% of the cases (28). Unfortunately, the authors did not conduct a direct comparison between sputum and plasma methylation data in their patient cohort. It is therefore not clear at the moment if the diagnostic efficacy could be further improved by analyzing plasma and sputum in parallel for the same patients.
In the described trial, the study population are those that had a suspicious lesion on screen CT and who underwent surgical resection, certainly a very select group and not necessarily generalizable to the population at risk. The control patients were from the same analysis, but not found to have cancer in the surgical specimen. Some of the control patients may have had lung cancer not yet discovered and the cancer patients may not necessarily represent a spectrum of biological disease that we currently see; lepidic-predominant disease, then called bronchioloalveolar, to invasive micropapillary with an aggressive histology. Lacking long-term follow-up information also makes it challenging to interpret the data. Finally, when combining an additional test with the CT, a clinician seeks a high negative predictive value, not necessarily a high sensitivity. Providing that information to the clinician will reduce unnecessary testing and reduce the burden of lung cancer screening.
Although Hulbert et al. did not notice a difference in methylation detection according to the size of the tumors and tumors of less than 2 cm diameter were readily scored, it is still likely that the location of the tumor within the bronchial tree may affect the analysis. Also, some patients may not be able to produce sputum, which necessitates analysis of plasma only. In summary, the new study reports promising data on methylation biomarkers that may aid in diagnosing patients with suspicious lesions found on CT scans. This type of analysis should now be extended to larger prospective studies in multi-center clinical trials.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Shaohua Cui (Department of Pulmonary Medicine, Shanghai Chest Hospital, Shanghai Jiao Tong University, Shanghai, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.01.17). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- National Lung Screening Trial Research Team. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011;365:395-409. [Crossref] [PubMed]
- Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev 2011;25:1010-22. [Crossref] [PubMed]
- Boulard M, Edwards JR, Bestor TH. FBXL10 protects Polycomb-bound genes from hypermethylation. Nat Genet 2015;47:479-85. [Crossref] [PubMed]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell 2007;128:683-92. [Crossref] [PubMed]
- Kalari S, Jung M, Kernstine KH, et al. The DNA methylation landscape of small cell lung cancer suggests a differentiation defect of neuroendocrine cells. Oncogene 2013;32:3559-68. [Crossref] [PubMed]
- Rauch T, Wang Z, Zhang X, et al. Homeobox gene methylation in lung cancer studied by genome-wide analysis with a microarray-based methylated CpG island recovery assay. Proc Natl Acad Sci U S A 2007;104:5527-32. [Crossref] [PubMed]
- Rauch TA, Zhong X, Wu X, et al. High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc Natl Acad Sci U S A 2008;105:252-7. [Crossref] [PubMed]
- Rauch TA, Wang Z, Wu X, et al. DNA methylation biomarkers for lung cancer. Tumour Biol 2012;33:287-96. [Crossref] [PubMed]
- Ohm JE, McGarvey KM, Yu X, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet 2007;39:237-42. [Crossref] [PubMed]
- Rauch T, Li H, Wu X, et al. MIRA-assisted microarray analysis, a new technology for the determination of DNA methylation patterns, identifies frequent methylation of homeodomain-containing genes in lung cancer cells. Cancer Res 2006;66:7939-47. [Crossref] [PubMed]
- Schlesinger Y, Straussman R, Keshet I, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet 2007;39:232-6. [Crossref] [PubMed]
- Widschwendter M, Fiegl H, Egle D, et al. Epigenetic stem cell signature in cancer. Nat Genet 2007;39:157-8. [Crossref] [PubMed]
- Shi H, Wang MX, Caldwell CW. CpG islands: their potential as biomarkers for cancer. Expert Rev Mol Diagn 2007;7:519-31. [Crossref] [PubMed]
- Perry AS, Watson RW, Lawler M, et al. The epigenome as a therapeutic target in prostate cancer. Nat Rev Urol 2010;7:668-80. [Crossref] [PubMed]
- Shivapurkar N, Gazdar AF. DNA methylation based biomarkers in non-invasive cancer screening. Curr Mol Med 2010;10:123-32. [Crossref] [PubMed]
- Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene 2002;21:5427-40. [Crossref] [PubMed]
- Herman JG, Baylin SB. Promoter-region hypermethylation and gene silencing in human cancer. Curr Top Microbiol Immunol 2000;249:35-54. [Crossref] [PubMed]
- Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer 2003;3:253-66. [Crossref] [PubMed]
- Ushijima T. Detection and interpretation of altered methylation patterns in cancer cells. Nat Rev Cancer 2005;5:223-31. [Crossref] [PubMed]
- Zöchbauer-Müller S, Minna JD, Gazdar AF. Aberrant DNA methylation in lung cancer: biological and clinical implications. Oncologist 2002;7:451-7. [Crossref] [PubMed]
- Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer 2004;4:988-93. [Crossref] [PubMed]
- Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet 2006;38:787-93. [Crossref] [PubMed]
- Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 2010;17:510-22. [Crossref] [PubMed]
- Fleischhacker M, Schmidt B. Circulating nucleic acids (CNAs) and cancer--a survey. Biochim Biophys Acta 2007;1775:181-232.
- Belinsky SA, Nikula KJ, Palmisano WA, et al. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci U S A 1998;95:11891-6. [Crossref] [PubMed]
- Esteller M, Sanchez-Cespedes M, Rosell R, et al. Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. Cancer Res 1999;59:67-70. [PubMed]
- Palmisano WA, Divine KK, Saccomanno G, et al. Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res 2000;60:5954-8. [PubMed]
- Hulbert A, Jusue Torres I, Stark A, et al. Early Detection of Lung Cancer using DNA Promoter Hypermethylation in Plasma and Sputum. Clin Cancer Res 2016; [Epub ahead of print]. [Crossref] [PubMed]
- Herman JG, Graff JR, Myöhänen S, et al. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A 1996;93:9821-6. [Crossref] [PubMed]
- Hubers AJ, Heideman DA, Burgers SA, et al. DNA hypermethylation analysis in sputum for the diagnosis of lung cancer: training validation set approach. Br J Cancer 2015;112:1105-13. [Crossref] [PubMed]


