
CRISPR made easy in human and murine hematopoietic precursors
The CRISPR/Cas9 technology has revolutionized gene-editing approaches, by favoring rapid and efficient modifications of gene function, from knock-out to knock-in, and even to silencing or activation of target genes (1,2). In the October 25th issue of Cell Reports, Gundry et al. (3) describe a novel and efficient method to engineer hematopoietic stem/progenitor cells (HSPCs), of both human and murine origin. In this study, highly efficient disruption of critical genes for myeloid cell development and function, including Dnmt3a, Eed and Suz12 (4), was achieved through virus-free systems. The authors achieve efficient delivery of small guide RNAs (sgRNAs) to murine Cas9-expressing HSPCs by electroporation, with subsequent knock-out of target genes, while maintaining cell viability and colony-forming properties of the edited cells. Targeting of Eed and Suz12 increased proliferation of the edited cells, and their capacity to serially replate, confirming the oncogenic properties of the gene targets. Importantly, the authors report complete virus-free delivery of both Cas9 and sgRNA in human primary T cells and in cord-blood derived HSPCs. By optimizing culture and transfection protocols they could achieve targeting efficiencies of over 80% in these cells. Off-target effects of the sgRNAs were detectable at a very low rate, while multilineage reconstitution capacity of stem cells was maintained.
One of the main advantages of the proposed method is the successful gene-editing of human HSPCs, not only by disruption, but through homology-directed repair (HDR), which allows introduction of point mutations at target loci, through co-transfection of a homologous “corrected” template (Figure 1). Strategies to increase HDR frequencies are widely studied (5,6) to improve current gene-editing strategies, which generally operate by inducing insertions or deletions (indels) through error-prone non-homologous end joining (NHEJ) of the target allele. To this end, introduction of precise genetic alterations through HDR in mammalian cells has the potential to be exploited for studies of cancer-associated point mutations or correction of disease-associated alterations in the clinical setting, extended, but not limited to, a large number of primary immunodeficiencies (7).
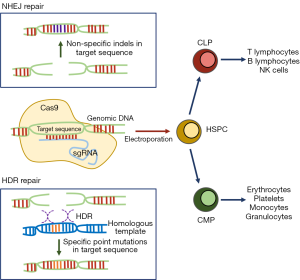
From a clinical standpoint, the virus-free nature of Gundry’s strategy represents a major advantage over a number of virus-based, generally retro or lentiviral based strategies (8), and their associated concerns, mainly related to insertional mutagenesis (9), and emergence of replication-competent viruses. Another important advantage of this technique is efficient co-transduction of combinations of Cas9 with multiple sgRNAs and repair templates to combine NHEJ- and HDR-mediated editing strategies. This particularly enables studies of loss-of-function and gain-of-function alterations together, which is highly relevant for cancer research but limited in systems based on constitutive Cas9 expression. It is easy to envision large applicability of this technology to studies of normal and malignant hematopoiesis, and to foresee applications in mouse modeling of myeloid and lymphoid malignancies, in line with previously reported CRISPR-based in vivo modeling of myeloid leukemias (10).
Overall, this technology represents a valuable and cost-effective resource, with large-scale applicability and accessibility, and an important tool to model genetic alterations in a wide variety of in vitro and in vivo settings.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Lorenzo Falchi (Division of Hematology/Oncology, Department of Medicine, Columbia University Medical Center, New York-Presbyterian Hospital, New York, USA).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.02.21). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Komor AC, Badran AH, Liu DR. CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell 2017;168:20-36. [Crossref] [PubMed]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014;157:1262-78. [Crossref] [PubMed]
- Gundry MC, Brunetti L, Lin A, et al. Highly Efficient Genome Editing of Murine and Human Hematopoietic Progenitor Cells by CRISPR/Cas9. Cell Rep 2016;17:1453-61. [Crossref] [PubMed]
- Shih AH, Abdel-Wahab O, Patel JP, et al. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer 2012;12:599-612. [Crossref] [PubMed]
- Richardson CD, Ray GJ, Bray NL, et al. Non-homologous DNA increases gene disruption efficiency by altering DNA repair outcomes. Nat Commun 2016;7:12463. [Crossref] [PubMed]
- Chu VT, Weber T, Wefers B, et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol 2015;33:543-8. [Crossref] [PubMed]
- Milner JD, Holland SM. The cup runneth over: lessons from the ever-expanding pool of primary immunodeficiency diseases. Nat Rev Immunol 2013;13:635-48. [Crossref] [PubMed]
- Kotterman MA, Chalberg TW, Schaffer DV. Viral Vectors for Gene Therapy: Translational and Clinical Outlook. Annu Rev Biomed Eng 2015;17:63-89. [Crossref] [PubMed]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 2003;302:415-9. [Crossref] [PubMed]
- Heckl D, Kowalczyk MS, Yudovich D, et al. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat Biotechnol 2014;32:941-6. [Crossref] [PubMed]


