
Mutations in KRAS: are they a valid biomarker for pancreatic ductal adenocarcinomas diagnosis?
Introduction
Of all cancers, pancreatic cancer is associated with the most detrimental clinical outcome. Even when treated according to most recent guidelines, more as ninety percent of patients will not survive the cancer beyond one to five years after diagnosis (1). Compounding the clinical problem posed by pancreatic cancer is its sheer size: no less as 337,872 cases were reported in 2012 and the number of cases is still increasing every year (2). A major factor in combating pancreatic cancer is the difficulty in identifying and differentiating the types of pancreatic cancers at an early stage. The present identification for this cancer is through computed tomography (CT), endoscopic retrograde cholangiopancreatogram (ERCP), Endoscopy ultrasound (EUS) followed with EUS-Fine needle aspiration biopsy (FNAB) and pathology (3), but optimism in this regard is growing due to the revolution in biomarker field per se. Molecular markers currently under assessment for their usefulness for the early diagnosis of pancreatic cancer include those based on the detection of genetic abnormalities within the mutational spectrum of pancreatic cancer, the detection of activated forms of signaling kinases involved in the progression of pancreatic cancer, the presence of specific miRNA variants in specific bodily fluids, distinct epigenetic alterations, as well as the finding of DNA with aberrant telomere length in fluids in contact with potential pancreatic cancers (4-6). The four major genes involved in the mutational spectrum for pancreatic ductal adenocarcinoma (PDAC) include P16, KRAS, TP53, and SMAD4 (1,7). Molecular markers involving detection of genetically aberrant variants of these genes or otherwise may aid making a definitive diagnosis of PDAC but may also guide gauging the potential response to chemotherapy and/or radiation therapy, thus improving success rates and help avoiding subjecting patients to side effect-prone treatment modalities (8). They may also guide assessing the extent of tumor heterogeneity during the course of cancer and thus the potential chances for success of treatment with targeted therapy like biological or novel kinase inhibitors (8). Sampling of material for determining biomarkers in suspected or established PDAC can be challenging: liquid biopsies (blood etc.), feces or saliva contain only minor amounts of tumor material, EUS-FNAB is invasive and produces also relatively little material and PDAC surgery is also challenging and the long time the procedure takes compromises quality of the biological material obtained. This situation is alleviated somewhat by the recent advent of technology capable of nanoscale DNA isolation and its analyses by modified new generation sequencing (NGS), digital PCR and QPCR while conversely better endoscopic procedures improve the quantity of material biopsy collected as well as the safety of the procedure involved. In conjunction these developments are starting to revolutionize the field but proof-of-principle studies are still relatively scarce, especially with regard to PDAC but also with cancerous disease in general (9-14). Important directions in the field aimed at providing such proof-of-principle for PDAC include nanoscale measurement of circulating or pancreatic cyst fluid cell free DNA (cfDNA), detection of circulating tumor cells (CTC) and analysis of exosomes from the blood. KRAS is ubiquitously mutated in PDAC and constitutes thus a rational target gene in this respect. Table 1 explains in detail of the different non-invasive methods that are under study and critical analysis involving KRAS and other genes which can be used as diagnostic and prognostic markers for PDAC.
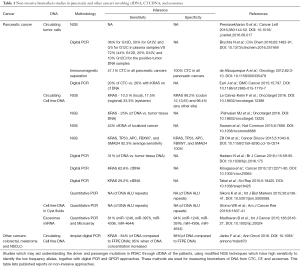
Full table
KRAS mutational spectrum in diagnosis of pancreatic cancer
An important advance in respect is that described by Le Calvez-Kelm et al. in a recent issue of oncotarget (15), who report on the use of KRAS mutation detection using as low as 2 ng of cfDNA with deep sequencing and Needlestack variant caller algorithm analysis from the patients with PDAC, chronic pancreatitis, and healthy controls. The study constitutes the largest screening of KRAS mutations in plasma samples of pancreatic cancer cases hitherto and its comparison to, other pathological pancreatic conditions and healthy controls allows for the comprehensive assessment of sensitivity and specificity of KRAS mutations using cfDNA. In this study the authors tried to understand how the KRAS mutations at the codon’s 12, 13 and 61 and new non-hotspot codons can affect the outcome of the disease as well to use it as the prospective diagnostic biomarker along in comparison to conventional plasma CA19.9 levels. Technically the study was exemplary as authors were able to identify with 0.08% of allele fraction detectability KRAS mutation status. Importantly, however, although the overall sensitivity in combination with CA19.9 increases to 95%, a limited sensitivity of 21.1% for the detection of pancreatic cancer was observed, substantially lower in comparison to CA19.9 alone. This would suggest that measuring cfDNA is not a promising avenue here and thus goes against the general direction of the field (see Table 1). The authors convincingly argue that other studies showing high specificity with KRAS did not include healthy controls (15). This method of specific amplicons obtained from a 2 ng DNA at the regions of exon2 and exon3 for the Kras hot spot codons are from exon 2 and exon 3 i.e. codon 12, codon 13 and codon 61, and non-hotspot codons 59, 62, 64 and 70 is an robust method to be used for other somatic mutations as in EGFR, TP53, SMAD4 and other DNA markers with in pancreatic cancer. Hence amplicon-based KRAS mutations sequencing is not as specific as anticipated, an important observation forcing the field to explore radically different avenues in this respect, e.g., microbiome determinations or mass spectrometry based directions.
Advantages and drawbacks of using KRAS and other mutational spectrum for diagnosis
Bailey et al., in the recent study have shown that the classification of different types of pancreatic cancer can be asserted using KRAS mutation and upregulation. The aberrantly differentiated endocrine exocrine origin PDAC’s carry the upregulated KRAS than other squamous, pancreatic progenitor and immunogenic tumors of pancreas (16). Kadayifci et al. showed the difference between the pancreatic cysts can be studied using DNA isolated from Cyst Fluids. They were able to obtain an overall accuracy of 86.2% when GNAS, KRAS and carcinoembryonic antigens determinations were combined, but this accuracy was markedly superior to when the individual determinations were used stand alone (17). In another study by Deshpande et al., the authors were able to identify a difference between pancreatic intra/epithelial tumors and the tumors driven into pancreatic duct from common bile duct origin using KRAS mutational analysis (95% vs. 11%) (18), thus although mutational analysis of cfDNA was very disappointing in the study Le Calvez-Kelm et al., analysis of mutation state per se will remain useful for diagnostic purposes when cellular material can be obtained.
Pancreatic cancers are among the most versatile forms of oncological disease. Preclinical work suggests that epithelial mesenchymal transition and the subsequent dissemination of tumor cells in the body is a very early event in this cancer, which already takes before obvious tumor formation and clear micrometastatic invasion into nearby tissues and lymph nodes can be detected (19). Encouragingly, work in colon cancer suggests that epithelial mesenchymal transition may be sensitive to various pharmacological inhibitors [e.g., ROCK inhibitors (20)] and thus very early detection through screening in combination with anti epithelial mesenchymal transition in high risk individuals remains an attractive proposition, despite the apparent difficulties in performing such screening through KRAS mutational analysis of cfDNA.
Nanoscale measurements of cfDNA and other DNA/RNA/proteins through non-invasive methods
Huang et al., showed that the use of AMP-based NGS methodology allows the detection of variants in allele frequencies as low as 1% and compared with allele-specific PCR as well digital PCR (1–5%) for KRAS, TP53, and SMAD4 in pancreatic cancer patients (21). There are other studies which may aid understanding the driver and passenger mutations in PDAC through cfDNA of the patients, using modified NGS techniques which have high sensitivity to identify the low frequency alleles, together with digital PCR and QPCR approaches. These methods are used for measuring biomarkers of DNA and RNA from CTC, CF, and exosomes (Tables 1,2). Table 2 gives the insights of the non-invasive non-DNA marker studies, which are aimed at improving pancreatic cancer diagnosis and prognosis. The same methods are being used in other cancers too like, colorectal, lung, liver and breast cancers in which such nanoscale measurements are showing some promise towards establishing cancer prognosis and may become useful for better therapy design. The study by Le Calvez-Kelm et al. shows that achieving this in PDAC may still entail a prolonged effort.

Full table
Conclusions
Pancreatic cancer with its associated infaust prognosis urgently needs better early detection of disease, especially for screening of high risk individuals. Despite early promise and theoretical considerations, however, KRAS mutional analysis of cfDNA seems not a way forward here. A more multifaceted panel of mutational spectrum biomarkers study (KRAS, TP53, SMAD4, P16, EGFR GNAS, MENN1, DAXX, VHL and in combination of pancreatic cancer specific STR markers) along with epigenetic alterations in cfDNA in pancreatic cancer patients may still provide a successful prognostic and diagnostic avenue, but generally speaking cfDNA does not appear very promising direction. Hence focus should now be directed to alternative methodology.
Acknowledgments
Funding: None.
Footnote
Correspondence to: Maikel P. Peppelenbosch. Erasmus MC Cancer Institute, Erasmus MC, Erasmus Medical Center Rotterdam, Na-1007, PO Box 2040, NL-3000 CA Rotterdam, the Netherlands. Email: M.Peppelenbosch@erasmusmc.nl.
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Xiaotian Sun (Department of Internal Medicine, Clinic of August First Film Studio, Beijing, China).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.02.46). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Rahib L, Fleshman JM, Matrisian LM, et al. Evaluation of Pancreatic Cancer Clinical Trials and Benchmarks for Clinically Meaningful Future Trials: A Systematic Review. JAMA Oncol 2016;2:1209-16. [Crossref] [PubMed]
- Ferlay J SI, Ervik M, Dikshit R, et al. GLOBOCAN 2012, IARC. Available online: http://globocan.iarc.fr/old/summary_table_site-html.asp?selection=23090&title=Pancreas&sex=0&type=0&window=1&africa=1&america=2&asia=3&europe=4&oceania=5&build=6&sort=0&submit=%C2%A0Execute
- Bruno MJ. Early diagnosis of pancreatic cancer; looking for a needle in a haystack? Gut 2013;62:955-6. [Crossref] [PubMed]
- Nuzhat Z, Kinhal V, Sharma S, et al. Tumour-derived exosomes as a signature of pancreatic cancer - liquid biopsies as indicators of tumour progression. Oncotarget 2016; [Epub ahead of print]. [PubMed]
- Herreros-Villanueva M, Bujanda L. Non-invasive biomarkers in pancreatic cancer diagnosis: what we need versus what we have. Ann Transl Med 2016;4:134. [Crossref] [PubMed]
- Utomo WK, Janmaat VT, Verhaar AP, et al. DNA integrity as biomarker in pancreatic cyst fluid. Am J Cancer Res 2016;6:1837-41. [PubMed]
- Braat H, Bruno M, Kuipers EJ, et al. Pancreatic cancer: promise for personalised medicine? Cancer Lett 2012;318:1-8. [Crossref] [PubMed]
- Goossens N, Nakagawa S, Sun X, et al. Cancer biomarker discovery and validation. Transl Cancer Res 2015;4:256-69. [PubMed]
- Teer JK. An improved understanding of cancer genomics through massively parallel sequencing. Transl Cancer Res 2014;3:243-59. [PubMed]
- Imamura T, Komatsu S, Ichikawa D, et al. Liquid biopsy in patients with pancreatic cancer: Circulating tumor cells and cell-free nucleic acids. World J Gastroenterol 2016;22:5627-41. [Crossref] [PubMed]
- Overbeek KA, Cahen DL, Canto MI, et al. Surveillance for neoplasia in the pancreas. Best Pract Res Clin Gastroenterol 2016;30:971-86. [Crossref] [PubMed]
- van Riet PA, Cahen DL, Poley JW, et al. Mapping international practice patterns in EUS-guided tissue sampling: outcome of a global survey. Endosc Int Open 2016;4:E360-70. [Crossref] [PubMed]
- Garcia-Murillas I, Schiavon G, Weigelt B, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med 2015;7:302ra133 [Crossref] [PubMed]
- Huang A, Zhang X, Zhou SL, et al. Detecting Circulating Tumor DNA in Hepatocellular Carcinoma Patients Using Droplet Digital PCR Is Feasible and Reflects Intratumoral Heterogeneity. J Cancer 2016;7:1907-14. [Crossref] [PubMed]
- Le Calvez-Kelm F, Foll M, Wozniak MB, et al. KRAS mutations in blood circulating cell-free DNA: a pancreatic cancer case-control. Oncotarget 2016;7:78827-40. [PubMed]
- Bailey P, Chang DK, Nones K, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531:47-52. [Crossref] [PubMed]
- Kadayifci A, Atar M, Wang JL, et al. Value of adding GNAS testing to pancreatic cyst fluid KRAS and carcinoembryonic antigen analysis for the diagnosis of intraductal papillary mucinous neoplasms. Dig Endosc 2017;29:111-7. [Crossref] [PubMed]
- Deshpande V, Konstantinidis IT, Castillo CF, et al. Intra-pancreatic Distal Bile Duct Carcinoma is Morphologically, Genetically, and Clinically Distinct from Pancreatic Ductal Adenocarcinoma. J Gastrointest Surg 2016;20:953-9. [Crossref] [PubMed]
- Rhim AD, Mirek ET, Aiello NM, et al. EMT and dissemination precede pancreatic tumor formation. Cell 2012;148:349-61. [Crossref] [PubMed]
- Voorneveld PW, Kodach LL, Jacobs RJ, et al. Loss of SMAD4 alters BMP signaling to promote colorectal cancer cell metastasis via activation of Rho and ROCK. Gastroenterology 2014;147:196-208.e13. [Crossref] [PubMed]
- Huang J, Löhr JM, Nilsson M, et al. Variant Profiling of Candidate Genes in Pancreatic Ductal Adenocarcinoma. Clin Chem 2015;61:1408-16. [Crossref] [PubMed]


