hangInhibition of the non homologous end joining process in the context of hypoxic tumor cells
Introduction
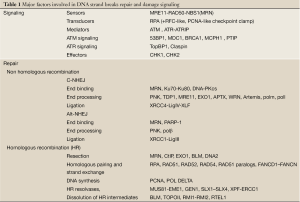
In addition to surgery, cancer treatment is based on radiotherapy and targeted/non-targeted chemotherapy or both. Radiotherapy as well as most of the non-targeted drugs provoke the formation of DNA damage including oxidized or alkylated nucleotides, single-strand breaks (SSBs) and double-strand breaks (DSBs). Amongst DNA damage, DSBs are considered as a highly toxic lesion that, if improperly repaired, can induce genomic rearrangement (1). Consequently the repair of DSBs has been subject to a lot of investigations. In mammalian cells, DSBs trigger the DNA damage response (DDR), an interconnected network that tends to maintain cell viability and genomic stability (2,3). The DDR acts through checkpoint signaling and DNA damage repair (4). In mammalian cells the MRE11/RAD50/NBS1 (MRN) complex binds to DSBs and facilitates the activation of Ataxia Telangiectasia Mutated (ATM), the key phosphatidylinositol 3-kinase protein kinase-like (PI3KK) in the DDR (5) (Table 1). At the break site ATM autophosphorylation allows its activation to subsequently phosphorylate a large number of substrates in the chromatin surrounding (6). In addition to ATM, two other members of the PI3KK family, ataxia telangiectasia and Rad3 related protein (ATR) and DNA-dependent protein kinase (DNA-PK) play prominent roles in the DDR through the detection of DNA replication fork collapse and DSBs repair, respectively.
Full table
DSBs repair is mainly dependent on the Non-homologous end joining (NHEJ) pathway that operates throughout the cell cycle (2,4,7-9). In contrast, the minor pathway, homologous recombination (HR), is active only during the S and G2 phases in which the sister chromatids are available to allow recombination processing. The balance between NHEJ and HR pathways is a prerequisite for efficient DSBs repair and relies on at least three factors: temporal control of proteins recruitment and modifications (10,11), chemical complexity of the breaks and chromatin conformation (12-14). Moreover a cross-talk between DNA-PKcs and ATM leads to a coordinated regulation of DSBs repair by NHEJ and HR (15).
Thus, the inhibition of DDR at the level of signaling and/or DNA damage repair by chemical compounds represents an area of research with the ultimate goal of sensitizing tumor cells to the treatment (16-19). Since the NHEJ pathway is under the control of DNA-PK, various compounds were selected to inhibit its activity. Interestingly, pharmaceutical companies did not take an interest in the research for DNA repair inhibitors since they favor the search for drugs that directly affect tumor cells or microenvironment. Recently, a renewed interest about DNA repair inhibitors was based on the killing effect of PARP inhibitors of BRCA1- and BRCA2-defective tumors (20-23). This example of an efficient monochemotherapy against solid tumors partially defective in DNA repair is reminiscent of the synthetic lethality mechanism (24-26).
Although DNA repair inhibitors are now in phase I/II trial, few of them being in phase III, one should keep in mind that tumor cells partly grow under hypoxia. The abnormal vasculature of tumors is considered as the most important contributor to the development of both chronic and acute hypoxia in the majority of solid tumors. Chronic hypoxia, or even anoxia in solid tumors is the consequence of abnormally long intravascular erythrocyte transit times. It is interesting to note that clinical evidence suggest that intra-tumoral hypoxia correlates with cell resistance to therapy as well as an aggressive behavior of the tumor leading to poor patient prognoses (27,28). A key regulator of the cellular response to hypoxia is the accumulation of the transcription factor hypoxia-inducible factor (HIF) which induces the expression of numerous target genes leading to cellular adaptation (29-31). Recent studies have reported diminished DNA repair capacities and increased mutagenesis in mammalian cells under hypoxic conditions (32-34). The NHEJ repair activity is affected in hypoxic cells and this short review summarizes the cross-talk between NHEJ and HIF pathways in the light of pharmacological inhibition of NHEJ in cancer therapy.
Hypoxic stress response and its impact on DNA repair
States of chronic hypoxia and transient hypoxia occur within solid tumors. Cells under chronic hypoxia are located beyond the diffusion limit of oxygen from a blood vessel (about 100 µm) whereas transient hypoxia corresponds to local oxygen depletion. Tumor adaptation to hypoxic stress occurs through modifications of its metabolism and induction of neovascularization. Oxygen concentrations of less than 0.02% (0.15 mmHg) render cells more resistant to killing by ionizing radiation by a factor of 2-3 (35). This stress response is controlled by HIF that binds to the cis-acting highly conserved consensus sequence 5'-G/ACGTG-3' referred as hypoxia response elements (HRE). The binding of HIF on HRE leads to increased expression of target genes involved in different pathways such as survival, glucose transport, glycolysis, angiogenesis, motility, basement membrane integrity and other functions (29,30,36). HIF is comprised of α- and β-subunits (37) with three HIFα isoforms reported to date, HIF-1α and HIF-2α being the best characterized. HIF-1α, the most ubiquitously expressed isoform, and HIF-2α regulate the expression of overlapping but non-identical genes (38,39). HIFα subunits are mainly targeted for normoxia-dependent degradation by the proteasomal system, whereas HIFβ subunits are constitutively expressed in most cells, HIF-1β/ARNT being the best characterized (29-31). Therefore, HIF activity is exquisitely dependent on the limiting expression of α subunit. Under normoxia HIFα subunits are hydroxylated by a family of prolyl hydroxylases (PHDs) (40-43) facilitating interaction with the von Hippel-Lindau (VHL) E3 ubiquitin ligase complex (44). Then, Hifα is targeted for ubiquitin-dependent degradation leading to a low level of protein content. Under hypoxic stress, the PHDs are inhibited leading to the stabilization of HIFα that translocates to the nucleus allowing its heterodimerization with HIF-1β/ARNT. The HIF heterodimer bound to HRE in an association with transcriptional coactivators as CBP/p300 activates a variety of hypoxia-responsive genes. HIF dependent transcriptional regulation contributes to the adaptive response to hypoxic conditions by upregulating more than 100 genes (45).
HIF-1 is overexpressed in many human cancers and correlates with poor prognosis outcome (29). This is consistent with studies demonstrating that tumor cells containing constitutively high levels of Hif1α were more resistant to both chemotherapy and radiotherapy (46-49). In most of these cases, overexpression is the consequence of a constitutive stabilization of the protein by hypoxia. However, there is increasing evidence demonstrating that a number of non-hypoxic stimuli such as genetic alterations that activate oncogenes and inactivate tumor suppressor genes or growth factors signaling are also highly capable of turning this transcription factor on (36). Accordingly, HIF-1 represents an attractive target for the development of pharmacological inhibitors (28,50,51).
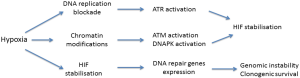
The impact of hypoxic stress has also been tested on DNA repair genes expression. After UVB irradiation, HIF-1 modulates the expression of XPC and XPD, two proteins of the nucleotide excision repair complex, with an increase of repair during the late phase of UV photoproduct removal (52). However, using an host-cell reactivation assay for repair of UV-damaged plasmid DNA, contradictory results of the effect of hypoxia on nucleotide excision repair were reported (53,54). Mismatch repair (MMR) which maintains genomic integrity by correcting replication errors is down-regulated under hypoxia through an epigenetic control (55-57). The consequence of this inhibitory effect is reminiscent to genomic instability in tumor cells grown under hypoxia. The recombination pathways are also affected by HIF-1 expression. HR repair proteins such as RAD51, BRCA2, and BRCA1 are compromised under hypoxic conditions (58-63). Contradictory results were reported on the expression of NHEJ proteins under hypoxia. Gene expression studies showed downregulation of mRNA encoding NHEJ proteins following chronic hypoxia but without change in protein content (62). In contrast under acute hypoxia an up-regulation of Ku70 expression and DNA-PKcs was reported as well as a direct interaction between DNA-PKcs and HIF1α (64,65). Very recently, an overexpresssion of Ku70/Ku80 proteins under the control of HIF-1 induction was reported (66). Then almost all of the DNA damage repair functions are repressed under hypoxia leading to cell sensitivity to therapy in contrast to the NHEJ pathway which is up-regulated (Figure 1). Under hypoxia, not only the expression of DNA-PK is enhanced but also its activity (67). DNA-PK is activated by mild hypoxia conditions (1% O2) while more severe hypoxia (0.1% O2) also activates ATM and ATR, two essential building blocks of the DNA DSBs signaling pathway. Hypoxia activates DNA-PK in the absence of DSBs in accordance with a study showing that activation of ATM in hypoxic cells arises independently of the MRN complex (68). Activation of DNA-PK or ATM could be triggered by stable association of single repair factors with chromatin in the absence of DNA lesions per se (69,70). Indeed, the DNA-PKcs autophosphorylation on S2056 was initiated by histone acetylation in response to hypoxia (67). These results illustrate the possibility of a non canonical DNA-PKcs activation which may be reminiscent to its activation under low salt conditions in the absence of Ku70/Ku80 (71).
NHEJ pathway and its pharmacological inhibition
Hypoxia can regulate DSBs repair through HR and NHEJ. In mammalian cells, NHEJ is the predominant repair pathway for DSBs which ligates the two DNA ends together with minimal end processing (72,73). NHEJ consists of at least two genetically and biochemically distinct sub-pathways: (I) a main canonical DNA-PK end-joining pathway named classical NHEJ (C-NHEJ) (7-9) and (II) an alternative or backup NHEJ end-joining pathway (Alt-NHEJ) (74-76).
C-NHEJ proceeds via at least three steps: (I) break recognition; (II) processing of the damaged DNA ends to remove non-ligatable groups; and (III) ligation to restore strand continuity. The prerequisite event for all the subsequent steps is the binding of the Ku70/Ku80 heterodimer to DNA ends (77). Live cell imaging studies following laser micro-irradiation indicate that core NHEJ components are independently recruited to Ku-bound DSBs (78), including the DNA-dependent protein kinase catalytic subunit (DNA-PKcs), XLF and the preassembled XRCC4/DNA Ligase IV complex (79). The DNA-PK holoenzyme is formed when DNA-PKcs binds to Ku at DSB ends and provides DNA ends recognition and protection activities followed by bridging the ends, associated with serine/threonine protein kinase activity (80). The kinase activity of DNA-PK is required for DSBs repair. In addition, DNA-PK conformational change mediated by autophosphorylation is necessary for the dissociation of the DNA-PKcs subunit from DNA ends and also for the activation of end-processing enzymes, such as the Artemis nuclease (81-83). Ligation requires the concerted action of LIG4, XRCC4 and XLF, the latter promoting re-adenylation of LIG4 (84,85). At a later stage in the C-NHEJ process, this molecular machinery is released from the re-ligated DNA.
Alt-NHEJ, detected only when C-NHEJ is compromised, exhibits a slower process (86) and produces deletions often accompanied by microhomology at the repair junction (76,87,88). This pathway relies on factors different from those involved in the C-NHEJ, such as poly (ADP-ribose) polymerase-1 (PARP-1), X-ray cross complementing factor 1 (XRCC1), DNA ligase III, polynucleotide kinase (PNK), or Flap endonuclease 1 (75,86,89-92). PARP-1 is a major sensor of DNA SSBs and participates to the core repair complex involving XRCC1-ligase III but also recognizes DSBs with very high affinity. Recently, the MRN complex has been implicated in the Alt-NEHJ mechanism (93-98). Alt-NHEJ is repressed under normal growing conditions by the Ku70/Ku80 heterodimer (86,99-104). This alternative NHEJ pathway may be particularly relevant to genomic instability associated with tumor development (105-108).
Since DDR defects are a common feature of tumor cells, the development of pharmacological inhibitors of DNA-PK, ATM and ATR, highlighted opportunities and challenges in cancer therapy (19,109). The core C-NHEJ complex was investigated as a target in relation with its involvement in radio- and chemoresistance of tumor cells (110,111). As soon as DNA-PK was described as the central partner of C-NHEJ (112-116), the search for specific inhibitors was undertaken (117). In addition, DNA-PK has been implicated in the repair of chlorambucil-induced cross-links, because increased DNA-PK activity in CLL cells correlates with clinical resistance to chlorambucil (118,119). Indeed, inhibitors of DNA-PKcs kinase activity have been shown to have efficacy in human cells (120,121) in line with the development of new compounds (109,122-124). In addition to these small chemical compounds, basically ATP-competitive inhibitors, antibody based inhibitors could also be effective as DNA-PK inhibitors. Recently, a DNA-PK specific antibody modified fragment ScFv18-2 was reported to decrease DNA-PK phosphorylation which correlates to radiosensitization (125). However, the therapeutic efficacy of targeting DNA-PK will depend in part on DNA-PK expression in tumor versus normal cells.
Among the different possibilities to inhibit NHEJ, the Ligase IV may be a better pharmacological target than DNA-PK itself, since the inhibition of the ligation step will not allow the Alt-NHEJ pathway to proceed due to the remaining Ku binding to DNA. Thus, the ligation complex was also investigated as an alternative strategy and inhibitors have been selected (126,127). However, the inhibitory effect should be complete since low levels of DNA ligases III and IV were sufficient for effective NHEJ which limits the potency of such alternative strategy (128). A different strategy is illustrated by the use of non specific kinase inhibitors such as KU-0060648 which is a potent dual inhibitor of DNA-PK and PI-3K. In relation with the role of the kinases in DSBs repair and the promotion of cell proliferation, KU-0060648 exhibits a direct effect but the toxicity is enhanced in cells treated with topoII inhibitors (120). At last, indirect approaches such as the use of short double-stranded DNA, Dbaits, have been shown to sensitize xenografted tumors to radiotherapy, not by inhibiting the kinase activity of DNA-PK, but by acting through the induction of “false” DNA damage signaling (129-131).
Whatever the NHEJ component selected as target, the inhibitors are mainly used as radio- or chemosensitizers in order to block DNA repair resulting in increased cell death. Such approaches enhance sensitivity to treatment, although they do not provide selectivity against cancer cells as they increase the radiosensitivity or chemosensitivity of normal cells as well. However, this drawback is related to chemotherapy since radiotherapy targets a defined tissue volume. Moreover, DDR defects are commonly associated to tumor cells with the loss of DNA repair processes resulting in genomic instability. Consequently such deficiency allows to potentially achieve selective antitumour activity through the inhibition of an essential DNA repair pathway such as NHEJ. The toxic activity of DDR inhibitors used in monochemotherapy was illustrated by the synthetic lethality obtained with PARP-1 inhibitors in BRCA1/BRCA2 deficient tumours which has raised the recent interest in the DDR modulation field. Despite a high level of expression of Ku and DNA-PKCS, an up-regulation of DNA-PKCS was reported in tumors or IR-resistant cell lines, suggesting a role in tumor growth and survival (132-135). Indeed, up-regulation of DNA-PK activity was shown to impair apoptosis in B-cell chronic lymphocytic leukemia (136). Moreover, in colorectal mismatch repair-deficient tumor cells, mutations in genes involved in DDR and DNA repair, including DNA-PKCS, have been reported (137). Taken together, all these alterations in DNA-PK expression or activity suggest that the consequences of its inhibition should be useful against tumors in line with a mechanism of toxicity based on the synthetic lethality process. However, in tumor tissues, the expression of DNA-PK shows intratumor heterogeneity, suggesting difficulty in predicting the radio- or chemo-sensitivity of the tumor as well as when a DNA-PK inhibitor might be beneficial (138).
HIF regulation by PI3KK recruited at DNA damage sites
HIF activity is regulated in two major ways. The first relies on hydroxylation-dependent degradation/inactivation of HIFα while the second involves oxygen-independent factors in a cell type-specific manner such as epidermal growth factor receptor (EGFR), heat-shock protein 90, phosphatidylinositol 3-kinase/AKT and MAPK, cyclooxygenase-2 activity (31,139-141). Interestingly, HIF-1α is also regulated by members of the PI3KK family, ATM, ATR and DNA-PK (Figure 1).
ATM protein deficiency correlates with an increased expression and activity of HIF-1α protein (142,143). Similarly, the inhibition of ATM in a mouse model suppressed the induction of senescence and leads to increased tumor size and invasiveness (144). Under hypoxia ATM phosphorylates HIF-1α at Ser696 site, which is required for its stability through putative posttranslational modifications such as sumoylation (143). Hereafter, ATM participates in conjunction with several other factors to the maintenance of an elevated mTORC1 activity in hypoxic tumors.
Hypoxia is known to induce a replication-associated damage response. For example, severe hypoxia induces S-phase arrest resulting in regions of single-stranded DNA at stalled replication forks and the activation of ATR (145). Loss of ATR results in a further loss of viability in S-phase cells under hypoxic conditions (146). Inhibition of ATR expression and activity inhibited cell survival upon hypoxic conditions (147). Under severe hypoxic conditions (0.1% O2), ATR is activated at early time points and this cellular stress response favors hypoxia adaptation by up-regulating the expression of both HIF-1α and HIF-1β/ARNT subunits, therefore contributing to cell adaptation to hypoxia (147). The mechanism of ATR dependent regulation of HIF-1α accumulation during hypoxia is at the stage of HIF-1α translation.
DNA-PK also participates to HIF regulation since the level of HIF-1α accumulation upon hypoxia is decreased in DNA-PK deficient cells (67). Similarly, the DNA-PK kinase activity, NU7026, provokes a decreased in HIF-1α content. Moreover, activated DNA-PK which is strictly dependent on Ku70/80 is able to regulate HIF-1α accumulation by interfering with the mechanisms protecting HIF-1α from nuclear degradation and subsequent increased expression of HIF-1 target genes (67).
Thus, the hypoxic accumulation of HIF-1 is positively regulated by ATM, ATR and DNA-PK that initiate cellular stress responses when either genome integrity, mRNA translation, or nutrient availability is compromised.
Conclusions and perspectives
The DDR (DNA damage response) induced during hypoxic stress has recently emerged as an important signaling pathway that allows cells to withstand modifications in environmental conditions. In contrast to other DNA-repair pathways, which were downregulated, leading to genetic instability (33), ATR, ATM and DNA-PK remained expressed and activated. Interestingly, the mechanism of hypoxia-induced activation of both DNA-PK and ATM is distinct from that of the DNA DSBs and stems on chromatin modifications. Taken together, activation of these three PI3KK during hypoxic stress allows HIF-1α accumulation in the nucleus. This regulation by non-hypoxic effectors influence HIF function, directly or indirectly, at different stages of its activation.Thus, the correlated regulation of PI3KKs with HIF-1 could contribute to therapy resistance in hypoxic tumor cells, and provides new evidence for developing therapeutic strategies enhancing the efficacy of cancer therapy in hypoxic tumor cells. In addition, hypoxia results in ATM-dependent phosphorylation of HIF-1α and mediates downregulation of mTORC1 signalling (143). Hereafter, inhibition of DNA-PK and ATM not only radiosensitize tumor cells but also interfere with HIF dependent regulation. Moreover, the use of NHEJ inhibitors alone could induce cell death through synthetic lethality, since DNA repair pathways and signaling are altered in tumors, most of them resulting of a mutated MMR pathway.
However, targeting the NHEJ pathway may be therapeutically inefficient or lead to unexpected results based, at least, on the following points:
(I) the dual effect on NHEJ and HIF activities presupposes drug delivery to the hypoxic region; (II) the expression of DNA-PK shows intratumor heterogeneity; (III) inhibition of DNA-PK may increase HR activity but also the Alt-NHEJ pathway; (IV) inhibition of DNA-PK is not expected to be specific towards tumor cells; (V) inhibition of DNA-PK may affect its function in regulatory mechanisms out of the canonical DSBs repair process (148). For instance DNA-PK plays a role in the USF-1-mediated transcriptional regulation of lipogenic genes during fasting/feeding (149). DNA-PK and Ku play other roles outside the nucleus (150) and may phosphorylate cytoplasmic targets involved in cellular signaling pathways (151). In addition, Ku has been reported as a moonlighting protein, displaying new functions in the cytoplasm or at the membrane level (152,153). Consequently, the potential effect of DNA-PK inhibitors with high or low specificity deserve to be investigated in vivo with tumor xenografts in order to take into account the complexity of these regulatory networks.
Acknowledgments
I acknowledge Drs P Calsou and C Muller from my previous team.
Funding: I acknowledge the financial support from ‘Ligue Nationale Contre le Cancer’ and Cancéropole GSO (Toulouse, France).
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (David J. Chen and Benjamin P.C. Chen) for the series “DNA Damage and Repair” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.3978/j.issn.2218-676X.2013.05.01). The series “DNA Damage and Repair” was commissioned by the editorial office without any funding or sponsorship. The author has no other conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- O’Driscoll M, Jeggo PA. The role of double-strand break repair - insights from human genetics. Nat Rev Genet 2006;7:45-54. [PubMed]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature 2009;461:1071-8. [PubMed]
- Kastan MB. DNA damage responses: mechanisms and roles in human disease: 2007 G.H.A. Clowes Memorial Award Lecture. Mol Cancer Res 2008;6:517-24. [PubMed]
- Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev 2011;25:409-33. [PubMed]
- Uziel T, Lerenthal Y, Moyal L, et al. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J 2003;22:5612-21. [PubMed]
- Kim YC, Gerlitz G, Furusawa T, et al. Activation of ATM depends on chromatin interactions occurring before induction of DNA damage. Nat Cell Biol 2009;11:92-6. [PubMed]
- Wang C, Lees-Miller SP. Detection and Repair of Ionizing Radiation-Induced DNA Double Strand Breaks: New Developments in Nonhomologous End Joining. Int J Radiat Oncol Biol Phys 2013;86:440-9. [PubMed]
- Weterings E, Chen DJ. The endless tale of non-homologous end-joining. Cell Res 2008;18:114-24. [PubMed]
- Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 2010;79:181-211. [PubMed]
- Chapman JR, Sossick AJ, Boulton SJ, et al. BRCA1-associated exclusion of 53BP1 from DNA damage sites underlies temporal control of DNA repair. J Cell Sci 2012;125:3529-34. [PubMed]
- Neal JA, Meek K. Choosing the right path: does DNA-PK help make the decision? Mutat Res 2011;711:73-86. [PubMed]
- Brandsma I, Gent DC. Pathway choice in DNA double strand break repair: observations of a balancing act. Genome Integr 2012;3:9. [PubMed]
- Kim JS, Krasieva TB, Kurumizaka H, et al. Independent and sequential recruitment of NHEJ and HR factors to DNA damage sites in mammalian cells. J Cell Biol 2005;170:341-7. [PubMed]
- Shibata A, Conrad S, Birraux J, et al. Factors determining DNA double-strand break repair pathway choice in G2 phase. EMBO J 2011;30:1079-92. [PubMed]
- Shrivastav M, Miller CA, De Haro LP, et al. DNA-PKcs and ATM co-regulate DNA double-strand break repair. DNA Repair (Amst) 2009;8:920-9. [PubMed]
- Basu B, Yap TA, Molife LR, et al. Targeting the DNA damage response in oncology: past, present and future perspectives. Curr Opin Oncol 2012;24:316-24. [PubMed]
- Begg AC, Stewart FA, Vens C. Strategies to improve radiotherapy with targeted drugs. Nat Rev Cancer 2011;11:239-53. [PubMed]
- Helleday T, Petermann E, Lundin C, et al. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer 2008;8:193-204. [PubMed]
- Finlay MR, Griffin RJ. Modulation of DNA repair by pharmacological inhibitors of the PIKK protein kinase family. Bioorg Med Chem Lett 2012;22:5352-9. [PubMed]
- Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005;434:913-7. [PubMed]
- Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005;434:917-21. [PubMed]
- Fong PC, Boss DS, Yap TA, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 2009;361:123-34. [PubMed]
- Fong PC, Yap TA, Boss DS, et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J Clin Oncol 2010;28:2512-9. [PubMed]
- Helleday T. Homologous recombination in cancer development, treatment and development of drug resistance. Carcinogenesis 2010;31:955-60. [PubMed]
- Chan N, Pires IM, Bencokova Z, et al. Contextual synthetic lethality of cancer cell kill based on the tumor microenvironment. Cancer Res 2010;70:8045-54. [PubMed]
- Helleday T. The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. Mol Oncol 2011;5:387-93. [PubMed]
- Chan N, Koch CJ, Bristow RG. Tumor hypoxia as a modifier of DNA strand break and cross-link repair. Curr Mol Med 2009;9:401-10. [PubMed]
- Monti E, Gariboldi MB. HIF-1 as a target for cancer chemotherapy, chemosensitization and chemoprevention. Curr Mol Pharmacol 2011;4:62-77. [PubMed]
- Brahimi-Horn MC, Pouysségur J. HIF at a glance. J Cell Sci 2009;122:1055-7. [PubMed]
- Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell 2010;40:294-309. [PubMed]
- Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer 2003;3:721-32. [PubMed]
- Chan N, Bristow RG. “Contextual” synthetic lethality and/or loss of heterozygosity: tumor hypoxia and modification of DNA repair. Clin Cancer Res 2010;16:4553-60. [PubMed]
- Bristow RG, Hill RP. Hypoxia and metabolism. Hypoxia, DNA repair and genetic instability. Nat Rev Cancer 2008;8:180-92. [PubMed]
- Bindra RS, Crosby ME, Glazer PM. Regulation of DNA repair in hypoxic cancer cells. Cancer Metastasis Rev 2007;26:249-60. [PubMed]
- Wouters BG, Brown JM. Cells at intermediate oxygen levels can be more important than the “hypoxic fraction” in determining tumor response to fractionated radiotherapy. Radiat Res 1997;147:541-50. [PubMed]
- Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell 2012;148:399-408. [PubMed]
- Wang GL, Jiang BH, Rue EA, et al. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A 1995;92:5510-4. [PubMed]
- Aprelikova O, Wood M, Tackett S, et al. Role of ETS transcription factors in the hypoxia-inducible factor-2 target gene selection. Cancer Res 2006;66:5641-7. [PubMed]
- Rankin EB, Biju MP, Liu Q, et al. Hypoxia-inducible factor-2 (HIF-2) regulates hepatic erythropoietin in vivo. J Clin Invest 2007;117:1068-77. [PubMed]
- Ivan M, Kondo K, Yang H, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 2001;292:464-8. [PubMed]
- Jaakkola P, Mole DR, Tian YM, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 2001;292:468-72. [PubMed]
- Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science 2001;294:1337-40. [PubMed]
- Epstein AC, Gleadle JM, McNeill LA, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 2001;107:43-54. [PubMed]
- Maxwell PH, Wiesener MS, Chang GW, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 1999;399:271-5. [PubMed]
- Xia X, Lemieux ME, Li W, et al. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc Natl Acad Sci U S A 2009;106:4260-5. [PubMed]
- Zhong H, De Marzo AM, Laughner E, et al. Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res 1999;59:5830-5. [PubMed]
- Unruh A, Ressel A, Mohamed HG, et al. The hypoxia-inducible factor-1 alpha is a negative factor for tumor therapy. Oncogene 2003;22:3213-20. [PubMed]
- Ryan HE, Poloni M, McNulty W, et al. Hypoxia-inducible factor-1alpha is a positive factor in solid tumor growth. Cancer Res 2000;60:4010-5. [PubMed]
- Aebersold DM, Burri P, Beer KT, et al. Expression of hypoxia-inducible factor-1alpha: a novel predictive and prognostic parameter in the radiotherapy of oropharyngeal cancer. Cancer Res 2001;61:2911-6. [PubMed]
- Semenza GL. Evaluation of HIF-1 inhibitors as anticancer agents. Drug Discov Today 2007;12:853-9. [PubMed]
- Wang Y, Liu Y, Malek SN, et al. Targeting HIF1α eliminates cancer stem cells in hematological malignancies. Cell Stem Cell 2011;8:399-411. [PubMed]
- Rezvani HR, Mahfouf W, Ali N, et al. Hypoxia-inducible factor-1alpha regulates the expression of nucleotide excision repair proteins in keratinocytes. Nucleic Acids Res 2010;38:797-809. [PubMed]
- Yuan J, Narayanan L, Rockwell S, et al. Diminished DNA repair and elevated mutagenesis in mammalian cells exposed to hypoxia and low pH. Cancer Res 2000;60:4372-6. [PubMed]
- Dregoesc D, Rainbow AJ. Differential effects of hypoxia and acidosis on p53 expression, repair of UVC-damaged DNA and viability after UVC in normal and tumor-derived human cells. DNA Repair (Amst) 2009;8:370-82. [PubMed]
- Rodríguez-Jiménez FJ, Moreno-Manzano V, Lucas-Dominguez R, et al. Hypoxia causes downregulation of mismatch repair system and genomic instability in stem cells. Stem Cells 2008;26:2052-62. [PubMed]
- Mihaylova VT, Bindra RS, Yuan J, et al. Decreased expression of the DNA mismatch repair gene Mlh1 under hypoxic stress in mammalian cells. Mol Cell Biol 2003;23:3265-73. [PubMed]
- Koshiji M, To KK, Hammer S, et al. HIF-1alpha induces genetic instability by transcriptionally downregulating MutSalpha expression. Mol Cell 2005;17:793-803. [PubMed]
- Bindra RS, Glazer PM. Repression of RAD51 gene expression by E2F4/p130 complexes in hypoxia. Oncogene 2007;26:2048-57. [PubMed]
- Bindra RS, Schaffer PJ, Meng A, et al. Down-regulation of Rad51 and decreased homologous recombination in hypoxic cancer cells. Mol Cell Biol 2004;24:8504-18. [PubMed]
- Chan N, Koritzinsky M, Zhao H, et al. Chronic hypoxia decreases synthesis of homologous recombination proteins to offset chemoresistance and radioresistance. Cancer Res 2008;68:605-14. [PubMed]
- Bindra RS, Gibson SL, Meng A, et al. Hypoxia-induced down-regulation of BRCA1 expression by E2Fs. Cancer Res 2005;65:11597-604. [PubMed]
- Meng AX, Jalali F, Cuddihy A, et al. Hypoxia down-regulates DNA double strand break repair gene expression in prostate cancer cells. Radiother Oncol 2005;76:168-76. [PubMed]
- Lu Y, Chu A, Turker MS, et al. Hypoxia-induced epigenetic regulation and silencing of the BRCA1 promoter. Mol Cell Biol 2011;31:3339-50. [PubMed]
- Kang MJ, Jung SM, Kim MJ, et al. DNA-dependent protein kinase is involved in heat shock protein-mediated accumulation of hypoxia-inducible factor-1alpha in hypoxic preconditioned HepG2 cells. FEBS J 2008;275:5969-81. [PubMed]
- Um JH, Kang CD, Bae JH, et al. Association of DNA-dependent protein kinase with hypoxia inducible factor-1 and its implication in resistance to anticancer drugs in hypoxic tumor cells. Exp Mol Med 2004;36:233-42. [PubMed]
- Ren Y, Hao P, Dutta B, et al. Hypoxia modulates A431 cellular pathways association to tumor radioresistance and enhanced migration revealed by comprehensive proteomic and functional studies. Mol Cell Proteomics 2013;12:485-98. [PubMed]
- Bouquet F, Ousset M, Biard D, et al. A DNA-dependent stress response involving DNA-PK occurs in hypoxic cells and contributes to cellular adaptation to hypoxia. J Cell Sci 2011;124:1943-51. [PubMed]
- Bencokova Z, Kaufmann MR, Pires IM, et al. ATM activation and signaling under hypoxic conditions. Mol Cell Biol 2009;29:526-37. [PubMed]
- Misteli T, Soutoglou E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol 2009;10:243-54. [PubMed]
- Soutoglou E, Misteli T. Activation of the cellular DNA damage response in the absence of DNA lesions. Science 2008;320:1507-10. [PubMed]
- Hammarsten O, Chu G. DNA-dependent protein kinase: DNA binding and activation in the absence of Ku. Proc Natl Acad Sci U S A 1998;95:525-30. [PubMed]
- Wyman C, Kanaar R. DNA double-strand break repair: all’s well that ends well. Annu Rev Genet 2006;40:363-83. [PubMed]
- Pardo B, Gómez-González B, Aguilera A. DNA repair in mammalian cells: DNA double-strand break repair: how to fix a broken relationship. Cell Mol Life Sci 2009;66:1039-56. [PubMed]
- Ma JL, Kim EM, Haber JE, et al. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol Cell Biol 2003;23:8820-8. [PubMed]
- Wang H, Perrault AR, Takeda Y, et al. Biochemical evidence for Ku-independent backup pathways of NHEJ. Nucleic Acids Res 2003;31:5377-88. [PubMed]
- Nussenzweig A, Nussenzweig MC. A backup DNA repair pathway moves to the forefront. Cell 2007;131:223-5. [PubMed]
- Downs JA, Jackson SP. A means to a DNA end: the many roles of Ku. Nat Rev Mol Cell Biol 2004;5:367-78. [PubMed]
- Yano K, Morotomi-Yano K, Wang SY, et al. Ku recruits XLF to DNA double-strand breaks. EMBO Rep 2008;9:91-6. [PubMed]
- Wu PY, Frit P, Meesala S, et al. Structural and functional interaction between the human DNA repair proteins DNA ligase IV and XRCC4. Mol Cell Biol 2009;29:3163-72. [PubMed]
- Meek K, Dang V, Lees-Miller SP. DNA-PK: the means to justify the ends? Adv Immunol 2008;99:33-58. [PubMed]
- Cui X, Yu Y, Gupta S, et al. Autophosphorylation of DNA-dependent protein kinase regulates DNA end processing and may also alter double-strand break repair pathway choice. Mol Cell Biol 2005;25:10842-52. [PubMed]
- Dobbs TA, Tainer JA, Lees-Miller SP. A structural model for regulation of NHEJ by DNA-PKcs autophosphorylation. DNA Repair (Amst) 2010;9:1307-14. [PubMed]
- Goodarzi AA, Yu Y, Riballo E, et al. DNA-PK autophosphorylation facilitates Artemis endonuclease activity. EMBO J 2006;25:3880-9. [PubMed]
- Riballo E, Woodbine L, Stiff T, et al. XLF-Cernunnos promotes DNA ligase IV-XRCC4 re-adenylation following ligation. Nucleic Acids Res 2009;37:482-92. [PubMed]
- Cottarel J, Frit P, Bombarde O, et al. A noncatalytic function of the ligation complex during nonhomologous end joining. J Cell Biol 2013;200:173-86. [PubMed]
- Wang M, Wu W, Wu W, et al. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res 2006;34:6170-82. [PubMed]
- Haber JE. Alternative endings. Proc Natl Acad Sci U S A 2008;105:405-6. [PubMed]
- McVey M, Lee SE. MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet 2008;24:529-38. [PubMed]
- Liang L, Deng L, Chen Y, et al. Modulation of DNA end joining by nuclear proteins. J Biol Chem 2005;280:31442-9. [PubMed]
- Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J Biol Chem 2004;279:55117-26. [PubMed]
- Audebert M, Salles B, Weinfeld M, et al. Involvement of polynucleotide kinase in a poly(ADP-ribose) polymerase-1-dependent DNA double-strand breaks rejoining pathway. J Mol Biol 2006;356:257-65. [PubMed]
- Wang H, Rosidi B, Perrault R, et al. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res 2005;65:4020-30. [PubMed]
- Deng Y, Guo X, Ferguson DO, et al. Multiple roles for MRE11 at uncapped telomeres. Nature 2009;460:914-8. [PubMed]
- Deriano L, Stracker TH, Baker A, et al. Roles for NBS1 in alternative nonhomologous end-joining of V(D)J recombination intermediates. Mol Cell 2009;34:13-25. [PubMed]
- Dinkelmann M, Spehalski E, Stoneham T, et al. Multiple functions of MRN in end-joining pathways during isotype class switching. Nat Struct Mol Biol 2009;16:808-13. [PubMed]
- Rass E, Grabarz A, Plo I, et al. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol 2009;16:819-24. [PubMed]
- Xie A, Kwok A, Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol 2009;16:814-8. [PubMed]
- Cheng Q, Barboule N, Frit P, et al. Ku counteracts mobilization of PARP1 and MRN in chromatin damaged with DNA double-strand breaks. Nucleic Acids Res 2011;39:9605-19. [PubMed]
- Weinstock DM, Brunet E, Jasin M. Formation of NHEJ-derived reciprocal chromosomal translocations does not require Ku70. Nat Cell Biol 2007;9:978-81. [PubMed]
- Fattah F, Lee EH, Weisensel N, et al. Ku regulates the non-homologous end joining pathway choice of DNA double-strand break repair in human somatic cells. PLoS Genet 2010;6:e1000855 [PubMed]
- Boboila C, Yan C, Wesemann DR, et al. Alternative end-joining catalyzes class switch recombination in the absence of both Ku70 and DNA ligase 4. J Exp Med 2010;207:417-27. [PubMed]
- Mansour WY, Schumacher S, Rosskopf R, et al. Hierarchy of nonhomologous end-joining, single-strand annealing and gene conversion at site-directed DNA double-strand breaks. Nucleic Acids Res 2008;36:4088-98. [PubMed]
- Guirouilh-Barbat J, Rass E, Plo I, et al. Defects in XRCC4 and KU80 differentially affect the joining of distal nonhomologous ends. Proc Natl Acad Sci U S A 2007;104:20902-7. [PubMed]
- Schulte-Uentrop L, El-Awady RA, Schliecker L, et al. Distinct roles of XRCC4 and Ku80 in non-homologous end-joining of endonuclease- and ionizing radiation-induced DNA double-strand breaks. Nucleic Acids Res 2008;36:2561-9. [PubMed]
- Boboila C, Jankovic M, Yan CT, et al. Alternative end-joining catalyzes robust IgH locus deletions and translocations in the combined absence of ligase 4 and Ku70. Proc Natl Acad Sci U S A 2010;107:3034-9. [PubMed]
- Rooney S, Chaudhuri J, Alt FW. The role of the non-homologous end-joining pathway in lymphocyte development. Immunol Rev 2004;200:115-31. [PubMed]
- Simsek D, Jasin M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nat Struct Mol Biol 2010;17:410-6. [PubMed]
- Soulas-Sprauel P, Rivera-Munoz P, Malivert L, et al. V(D)J and immunoglobulin class switch recombinations: a paradigm to study the regulation of DNA end-joining. Oncogene 2007;26:7780-91. [PubMed]
- Davidson D, Amrein L, Panasci L, et al. Small Molecules, Inhibitors of DNA-PK, Targeting DNA Repair, and Beyond. Front Pharmacol 2013;4:5. [PubMed]
- Salles B, Calsou P, Frit P, et al. The DNA repair complex DNA-PK, a pharmacological target in cancer chemotherapy and radiotherapy. Pathol Biol (Paris) 2006;54:185-93. [PubMed]
- Collis SJ, DeWeese TL, Jeggo PA, et al. The life and death of DNA-PK. Oncogene 2005;24:949-61. [PubMed]
- Blunt T, Finnie NJ, Taccioli GE, et al. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell 1995;80:813-23. [PubMed]
- Gottlieb TM, Jackson SP. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell 1993;72:131-42. [PubMed]
- Hartley KO, Gell D, Smith GC, et al. DNA-dependent protein kinase catalytic subunit: a relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell 1995;82:849-56. [PubMed]
- Lees-Miller SP, Chen YR, Anderson CW. Human cells contain a DNA-activated protein kinase that phosphorylates simian virus 40 T antigen, mouse p53, and the human Ku autoantigen. Mol Cell Biol 1990;10:6472-81. [PubMed]
- Lees-Miller SP, Godbout R, Chan DW, et al. Absence of p350 subunit of DNA-activated protein kinase from a radiosensitive human cell line. Science 1995;267:1183-5. [PubMed]
- Take Y, Kumano M, Hamano Y, et al. OK-1035, a selective inhibitor of DNA-dependent protein kinase. Biochem Biophys Res Commun 1995;215:41-7. [PubMed]
- Christodoulopoulos G, Muller C, Salles B, et al. Potentiation of chlorambucil cytotoxicity in B-cell chronic lymphocytic leukemia by inhibition of DNA-dependent protein kinase activity using wortmannin. Cancer Res 1998;58:1789-92. [PubMed]
- Muller C, Christodoulopoulos G, Salles B, et al. DNA-Dependent protein kinase activity correlates with clinical and in vitro sensitivity of chronic lymphocytic leukemia lymphocytes to nitrogen mustards. Blood 1998;92:2213-9. [PubMed]
- Munck JM, Batey MA, Zhao Y, et al. Chemosensitization of cancer cells by KU-0060648, a dual inhibitor of DNA-PK and PI-3K. Mol Cancer Ther 2012;11:1789-98. [PubMed]
- Sun X, Yang C, Liu H, et al. Identification and characterization of a small inhibitory peptide that can target DNA-PKcs autophosphorylation and increase tumor radiosensitivity. Int J Radiat Oncol Biol Phys 2012;84:1212-9. [PubMed]
- Cano C, Barbeau OR, Bailey C, et al. DNA-dependent protein kinase (DNA-PK) inhibitors. Synthesis and biological activity of quinolin-4-one and pyridopyrimidin-4-one surrogates for the chromen-4-one chemotype. J Med Chem 2010;53:8498-507. [PubMed]
- Clapham KM, Bardos J, Finlay MR, et al. DNA-dependent protein kinase (DNA-PK) inhibitors: structure-activity relationships for O-alkoxyphenylchromen-4-one probes of the ATP-binding domain. Bioorg Med Chem Lett 2011;21:966-70. [PubMed]
- Maira SM, Pecchi S, Huang A, et al. Identification and characterization of NVP-BKM120, an orally available pan-class I PI3-kinase inhibitor. Mol Cancer Ther 2012;11:317-28. [PubMed]
- Xiong H, Lee RJ, Haura EB, et al. Intranuclear delivery of a novel antibody-derived radiosensitizer targeting the DNA-dependent protein kinase catalytic subunit. Int J Radiat Oncol Biol Phys 2012;83:1023-30. [PubMed]
- Srivastava M, Nambiar M, Sharma S, et al. An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell 2012;151:1474-87. [PubMed]
- Chen X, Zhong S, Zhu X, et al. Rational design of human DNA ligase inhibitors that target cellular DNA replication and repair. Cancer Res 2008;68:3169-77. [PubMed]
- Windhofer F, Wu W, Iliakis G. Low levels of DNA ligases III and IV sufficient for effective NHEJ. J Cell Physiol 2007;213:475-83. [PubMed]
- Berthault N, Maury B, Agrario C, et al. Comparison of distribution and activity of nanoparticles with short interfering DNA (Dbait) in various living systems. Cancer Gene Ther 2011;18:695-706. [PubMed]
- Dutreix M, Cosset JM, Sun JS. Molecular therapy in support to radiotherapy. Mutat Res 2010;704:182-9. [PubMed]
- Quanz M, Berthault N, Roulin C, et al. Small-molecule drugs mimicking DNA damage: a new strategy for sensitizing tumors to radiotherapy. Clin Cancer Res 2009;15:1308-16. [PubMed]
- Ader I, Muller C, Bonnet J, et al. The radioprotective effect of the 24 kDa FGF-2 isoform in HeLa cells is related to an increased expression and activity of the DNA dependent protein kinase (DNA-PK) catalytic subunit. Oncogene 2002;21:6471-9. [PubMed]
- Beskow C, Skikuniene J, Holgersson A, et al. Radioresistant cervical cancer shows upregulation of the NHEJ proteins DNA-PKcs, Ku70 and Ku86. Br J Cancer 2009;101:816-21. [PubMed]
- Hosoi Y, Watanabe T, Nakagawa K, et al. Up-regulation of DNA-dependent protein kinase activity and Sp1 in colorectal cancer. Int J Oncol 2004;25:461-8. [PubMed]
- Söderlund Leifler K, Queseth S, Fornander T, et al. Low expression of Ku70/80, but high expression of DNA-PKcs, predict good response to radiotherapy in early breast cancer. Int J Oncol 2010;37:1547-54. [PubMed]
- Deriano L, Guipaud O, Merle-Béral H, et al. Human chronic lymphocytic leukemia B cells can escape DNA damage-induced apoptosis through the nonhomologous end-joining DNA repair pathway. Blood 2005;105:4776-83. [PubMed]
- Miquel C, Jacob S, Grandjouan S, et al. Frequent alteration of DNA damage signalling and repair pathways in human colorectal cancers with microsatellite instability. Oncogene 2007;26:5919-26. [PubMed]
- Tonotsuka N, Hosoi Y, Miyazaki S, et al. Heterogeneous expression of DNA-dependent protein kinase in esophageal cancer and normal epithelium. Int J Mol Med 2006;18:441-7. [PubMed]
- Hudson CC, Liu M, Chiang GG, et al. Regulation of hypoxia-inducible factor 1alpha expression and function by the mammalian target of rapamycin. Mol Cell Biol 2002;22:7004-14. [PubMed]
- Lee BL, Kim WH, Jung J, et al. A hypoxia-independent up-regulation of hypoxia-inducible factor-1 by AKT contributes to angiogenesis in human gastric cancer. Carcinogenesis 2008;29:44-51. [PubMed]
- Chun SY, Johnson C, Washburn JG, et al. Oncogenic KRAS modulates mitochondrial metabolism in human colon cancer cells by inducing HIF-1α and HIF-2α target genes. Mol Cancer 2010;9:293. [PubMed]
- Ousset M, Bouquet F, Fallone F, et al. Loss of ATM positively regulates the expression of hypoxia inducible factor 1 (HIF-1) through oxidative stress: Role in the physiopathology of the disease. Cell Cycle 2010;9:2814-22. [PubMed]
- Cam H, Easton JB, High A, et al. mTORC1 signaling under hypoxic conditions is controlled by ATM-dependent phosphorylation of HIF-1α. Mol Cell 2010;40:509-20. [PubMed]
- Bartkova J, Rezaei N, Liontos M, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 2006;444:633-7. [PubMed]
- Hammond EM, Dorie MJ, Giaccia AJ. ATR/ATM targets are phosphorylated by ATR in response to hypoxia and ATM in response to reoxygenation. J Biol Chem 2003;278:12207-13. [PubMed]
- Hammond EM, Dorie MJ, Giaccia AJ. Inhibition of ATR leads to increased sensitivity to hypoxia/reoxygenation. Cancer Res 2004;64:6556-62. [PubMed]
- Fallone F, Britton S, Nieto L, et al. ATR controls cellular adaptation to hypoxia through positive regulation of hypoxia-inducible factor 1 (HIF-1) expression. Oncogene 2013;32:4387-96. [PubMed]
- Kong X, Shen Y, Jiang N, et al. Emerging roles of DNA-PK besides DNA repair. Cell Signal 2011;23:1273-80. [PubMed]
- Wong RH, Chang I, Hudak CS, et al. A role of DNA-PK for the metabolic gene regulation in response to insulin. Cell 2009;136:1056-72. [PubMed]
- Huston E, Lynch MJ, Mohamed A, et al. EPAC and PKA allow cAMP dual control over DNA-PK nuclear translocation. Proc Natl Acad Sci U S A 2008;105:12791-6. [PubMed]
- Quanz M, Herbette A, Sayarath M, et al. Heat shock protein 90α (Hsp90α) is phosphorylated in response to DNA damage and accumulates in repair foci. J Biol Chem 2012;287:8803-15. [PubMed]
- Muller C, Paupert J, Monferran S, et al. The double life of the Ku protein: facing the DNA breaks and the extracellular environment. Cell Cycle 2005;4:438-41. [PubMed]
- Monferran S, Paupert J, Dauvillier S, et al. The membrane form of the DNA repair protein Ku interacts at the cell surface with metalloproteinase 9. EMBO J 2004;23:3758-68. [PubMed]