Translating prognostic prostate cancer gene signatures into the clinic
Traditional prognostic factors PSA, Gleason score and TNM staging allow patients with prostate cancer to be classified into low, intermediate and high risk groups (1). These prognostic groups are used not only to predict clinical outcome, but also to discuss appropriate treatment options with patients. Recently the Gleason score was further refined into the Gleason Group Grade to reflect the differences in clinical outcome between Gl 3+4 and Gl 4+3 (2).
The landmark ProTect study published last year (3) confirmed what clinicians had known for some time, that low and intermediate risk prostate cancer can be managed conservatively with excellent outcomes. For those who go on to have radical treatment surgery and radiotherapy are equally effective for cancer outcomes although long term side-effects are more marked with surgery. The outcomes for low and intermediate risk prostate cancer are excellent, but 10% of patients treated with state-of-the-art radiotherapy will experience biochemical relapse (4,5). For men with high-risk, locally advanced prostate cancer, 3-year recurrence rates can be in the order of 30% (5) and the 5-year failure-free survival rate for those with metastatic disease is around 30% (6). However, there is a spectrum of response even within the defined risk groups. Clearly there are limitations to the current risk classification groups, and underpinning this variation in clinical outcome is the genetic heterogeneity of prostate cancer. Analysis of 4,938,362 mutations from 7,042 cancers showed the diversity of mutational processes underlying the development of cancer. The prevalence of somatic mutations was highly variable between tumor types being high in melanoma low in acute lymphocytic leukemia and intermediate in prostate cancer (7). The level of genomic alterations was also highly variable within each tumor type.
In recent years there has been a plethora of advances in molecular technology. It is now easier and cheaper than ever to interrogate the cancer genome. Although associations have been found between individual genes as well as gene panels, there are no robustly validated genetic biomarkers which are used routinely in clinical practice (8). A number of commercially available products show potential, but there is still some way to go before there are enough data to convincingly demonstrate added value to prostate cancer patients. The commercially available RNA based signatures for prostate cancer are the 22-gene Decipher assay that is prognostic for risk of metastasis following prostatectomy; the 31-gene Prolaris test (46 genes including internal reference genes) that assesses aggressiveness; and the 17-gene Oncotype DX Prostate Score that tests the probability of metastatic disease (9).
There is now a DNA classifier to add to the validated prognostic RNA based signatures. In the November 2016 on line issue of European Urology, Lalonde et al. (10) presented work aimed at validating a prognostic DNA genomic classifier and progressing its translation into an aid to guide treatment planning. In an earlier publication, the collaborative group derived a DNA-based 100-locus copy number alteration (CNA) genomic classifier that stratified localized prostate cancers into groups with low and high risks of recurrence (11). In the European Oncology paper, the classifier was reduced to a 31-locus test by evaluating changes in RNA levels. Loci were selected where RNA expression reflected the copy number state. Thirty-one loci were identified that involved 109 genes, and the reduced signature was validated in four retrospective cohorts totalling 563 radical prostatectomy patients. The 31-locus genomic classifier identified patients with an increased risk of biochemical relapse [hazard ratio (HR) =2.73, P<0.001] and risk of metastasis (HR =7.79, P<0.001). Combining the classifier with standard prognostic variables outperformed use of clinical models alone. A further cohort of 102 patients was used to measure and validate the 31-locus classifier using the NanoString platform, which is suitable for clinical application. The 100-locus genomic classifier was shown in an earlier publication to outperform published RNA signatures including the OncoType DX Genomic Prostate Score and Prolaris test (11).
Precision medicine initiatives are striving to optimize therapies for patient sub-groups based on genetic or molecular profiling. The current most promising prostate cancer signatures have been validated in terms of prognostication to justify their use to aid decisions of whether to treat or to intensify treatment. They have not, however, been evaluated prospectively to show they improve outcomes or can predict benefit from specific interventions. Lalonde et al. highlight this limitation and state that future prospective trials will need “to evaluate whether the genomic classifier can serve as a predictive biomarker”, i.e., show that treatment intensification improves outcomes. The use of a standardized NanoString platform will aid future prospective validation.
The design of a follow-on interventional trial requires consideration of the choice of appropriate treatment. In a low risk group it would be important to select the patients who are not suitable for active surveillance so that either surgery or radiotherapy could be discussed. Where radiotherapy is the treatment of choice decisions regarding dose escalation or de-escalation could permit tailored treatment optimizing both cancer outcome and long-term toxicity risk. Defining groups of patients who would benefit from combined androgen deprivation therapy (ADT) and radiotherapy as well as those who would benefit from longer term ADT would allow targeting of treatment that can be beneficial, but can also have significant effects on a man’s quality-of-life.
One of the most important questions in radiotherapy is when to irradiate the pelvic nodes for patient benefit, and prognostic stratification could select a group for which lymph node irradiation increases cure. For patients with high risk prostate cancer, the STAMPEDE trial has shown the benefit of early chemotherapy (12). However, there is little doubt that chemotherapy is toxic and for some patients can adversely affect their quality-of-life so any steer towards selecting patients who benefit would be welcome.
The use of genomic signatures to improve prognostication would be a game-changer, but even more exciting would be the use of genetic indicators to predict specific treatment benefit. Connectivity mapping has been used to identify link RNA signatures with novel or re-purposed drugs which may be used to enhance treatment (13). Future research could use a network of genes based on the transcriptomic signature associated with the genomic classifier, and then use connectivity mapping to identify possible FDA approved agents for re-purposing. An ultimate goal for a radiotherapy-predictive biomarker would be to stratify patients who benefit from different modes of radiotherapy: low dose-rate brachytherapy, high dose-rate brachytherapy, protons or photons. Research aimed towards the latter requires generation of cohorts reflecting the different types of radiotherapy. As almost all signature generation to date has involved surgical cohorts and given the importance of radiotherapy in the treatment of the disease, there is a clear need to collect radiotherapy cohorts.
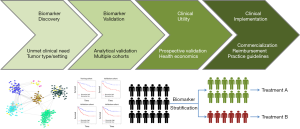
The speed of technological development highlights the challenges faced in translating gene signatures into the clinic. Biomarker discovery is easy but it is much harder to obtain the funding for qualifying a biomarker for clinical use. Tests need to be validated analytically and clinically and then shown to have clinical utility and an ability to improve healthcare (Figure 1). It is a highly competitive field that requires multi-disciplinary expertise and multi-center collaboration. The paper by Lalonde et al. illustrate the depth and breadth of research required. The work also illustrates the potential. However, it is a competitive field and the need to show clinical utility is paramount within an increasingly crowded area.
Acknowledgments
Funding: The authors are supported by Prostate Cancer UK and Cancer Research UK,, and acknowledge funding received by Prostate Cancer UK (PG14-008-TR2) and the Belfast Manchester Movember Centre of Excellence (CEO13_2_004).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Peng Zhang (Department of Urology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.03.13). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- D'Amico AV, Whittington R, Malkowicz SB, et al. Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. JAMA 1998;280:969-74. [Crossref] [PubMed]
- Epstein JI, Zelefsky MJ, Sjoberg DD, et al. A Contemporary Prostate Cancer Grading System: A Validated Alternative to the Gleason Score. Eur Urol 2016;69:428-35. [Crossref] [PubMed]
- Donovan JL, Hamdy FC, Lane JA, et al. Patient-Reported Outcomes after Monitoring, Surgery, or Radiotherapy for Prostate Cancer. N Engl J Med 2016;375:1425-37. [Crossref] [PubMed]
- Dearnaley D, Syndikus I, Mossop H, et al. Conventional versus hypofractionated high-dose intensity-modulated radiotherapy for prostate cancer: 5-year outcomes of the randomised, non-inferiority, phase 3 CHHiP trial. Lancet Oncol 2016;17:1047-60. [Crossref] [PubMed]
- James ND, Spears MR, Clarke NW, et al. Failure-Free Survival and Radiotherapy in Patients With Newly Diagnosed Nonmetastatic Prostate Cancer: Data From Patients in the Control Arm of the STAMPEDE Trial. JAMA Oncol 2016;2:348-57. [Crossref] [PubMed]
- James ND, Sydes MR, Clarke NW, et al. Addition of docetaxel, zoledronic acid, or both to first-line long-term hormone therapy in prostate cancer (STAMPEDE): survival results from an adaptive, multiarm, multistage, platform randomised controlled trial. Lancet 2016;387:1163-77. [Crossref] [PubMed]
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415-21. [Crossref] [PubMed]
- Boström PJ, Bjartell AS, Catto JW, et al. Genomic Predictors of Outcome in Prostate Cancer. Eur Urol 2015;68:1033-44. [Crossref] [PubMed]
- Nguyen HG, Welty CJ, Cooperberg MR. Diagnostic associations of gene expression signatures in prostate cancer tissue. Curr Opin Urol 2015;25:65-70. [Crossref] [PubMed]
- Lalonde E, Alkallas R, Chua ML, et al. Translating a Prognostic DNA Genomic Classifier into the Clinic: Retrospective Validation in 563 Localized Prostate Tumors. Eur Urol 2016; [Epub ahead of print]. [Crossref] [PubMed]
- Lalonde E, Ishkanian AS, Sykes J, et al. Tumour genomic and microenvironmental heterogeneity for integrated prediction of 5-year biochemical recurrence of prostate cancer: a retrospective cohort study. Lancet Oncol 2014;15:1521-32. [Crossref] [PubMed]
- Vale CL, Burdett S, Rydzewska LH, et al. Addition of docetaxel or bisphosphonates to standard of care in men with localised or metastatic, hormone-sensitive prostate cancer: a systematic review and meta-analyses of aggregate data. Lancet Oncol 2016;17:243-56. [Crossref] [PubMed]
- McArt DG, Dunne PD, Blayney JK, et al. Connectivity Mapping for Candidate Therapeutics Identification Using Next Generation Sequencing RNA-Seq Data. PloS One 2013;8:e66902 [Crossref] [PubMed]