
The subtype-specific role of histone H1.0 in cancer cell differentiation and intratumor heterogeneity
Cancer originates in a single cell. However, most human tumors present extensive intratumor heterogeneity and comprise several populations with distinct phenotypes. Intratumor heterogeneity have to be considered in the design of treatment strategies. Torres et al. (1) has identified an epigenetic mechanism that creates intratumor functional heterogeneity. They report that reversible silencing of histone H1 subtype H1.0 affects the differentiation state of cancer cells.
Histone H1 is involved in chromatin organization and gene regulation. In humans, the H1 complement is formed by 11 different subtypes, which include seven somatic subtypes and four germ-line-specfic subtypes (2). Somatic subtypes are classified as replication-dependent subtypes (H1.1-H1.5) and replication-independent subtypes (H1.0 and H1.x). While H1 subtypes display some degree of functional redundancy, increasing evidence is in favor of subtype functional differentiation. Subtypes differ in several aspects including expression patterns, chromatin binding affinity, post-translational modifications, protein-protein interactions and genomic distribution (3).
For more than 30 years histone H1.0 accumulation has been associated with terminal differentiation in normal cells (4,5), while a decrease in the levels of H1.0 expression has been observed in neoplastic cells (6). During the last decade, the down-regulation of H1.0 has been consistently observed in different cancer types (1,7-11). Recently in Science, Torres and colleagues have shown that H1.0 silencing occurs specifically in cancer stem cells (CSCs). They studied tumor formation using de novo transformed primary dermal fibroblasts. After transformation, a subpopulation of cells acquired a cancer-stem-cell phenotype, characterized by uncontrolled long-term proliferative potential and pluripotency. At a molecular level, CSCs are characterized by the expression of the stem cell surface marker Stage-Specific Embryonic Antigen 1 (SSEA1), which showed a mutually exclusive relationship with H1.0. At protein level, down-regulation of H1.0 could either decrease the total H1:core histone ratio or instead H1.0 loss could be compensated by an increase of other H1 subtypes. In this context, previous results have shown the increase of the protein levels of other H1 subtypes in different cancers (10).
Previous studies have reported the localization of H1.0 at genome-wide level. In mouse embryonic stem cells (ESCs), overexpression of H1.0 showed an overall positive correlation with H3K9me3 and inverse correlations with GC% and H3K4me3. H1.0 also appeared to be enriched at major and minor satellites and, to a lesser extent, at LINE L1 elements (12). The knockdown of H1.0, during in vitro differentiation experiments of human ESCs, impaired the induction of genes associated with differentiation (HNF4, Sox17 and FoxA2), suggesting a potential role of H1.0 in regulating specific genes during human ESC differentiation (13). In T47D, a human breast cancer cell-line, H1.0 was enriched at nucleolar-associated domains (NADs), rDNA and certain repeats (14).
An obvious question arising from this data regards the alterations derived from H1.0 silencing in CSCs, Torres et al. examined the genomic distribution of H1.0 in vitro transformed fibroblasts. H1.0 was preferentially located in GC-rich regions, while AT-rich regions were characterized by a low H1.0 peak density. The analysis of transcriptional data showed that the genes up-regulated upon H1.0 knockdown were associated with oncogene activation, tumor-suppressor inactivation and stem-cell maintenance, highlighting their importance for cancer cell-renewal ability. Additionally, the up-regulated genes were located in AT-rich regions with linear proximity on the chromosome. ChIP-seq analysis showed that the up-regulated genes were enriched in H1.0 around the transcriptional start site (TSS). Depletion of H1.0 caused an increase in the FAIRE signal around the TSS, indicating decreased nucleosome occupancy. Based on these results the authors concluded that cells lacking H1.0 undergo genome wide changes in gene expression, triggered by alterations in the nucleosome occupancy around the TSS of genes in AT-rich domains, which led to the up-regulation of genes associated with cancer-cell self-renewal ability. Mechanistically, H1.0 depletion destabilizes nucleosome-DNA interactions to promote the expression of self-renewal genes. Gene expression changes following H1.0 loss were reversible and epigenetic states restricting cell proliferation potential were reestablished upon H1.0 re-expression.
Torres et al. examined clinical samples from different human cancer types and confirmed that the decrease in the levels of H1.0 is a common feature of neoplastic cells. Moreover, low H1.0 levels predicted poor patient prognosis, underscoring the potential of this linker histone subtype as a cancer biomarker. Of special interest for therapeutic purposes is the fact that re-expression of H1.0 during tumor growth contributes to cell differentiation and impairs cell long-term proliferative potential.
In order to understand how H1.0 is silenced in CSCs, the authors studied the methylation levels of the H1.0 gene (H1F0). The comparison of two populations, self-renewing cells (SSEA1+) with the more differentiated tumor cells (SSEA1−) showed that the CpG island in the H1F0 promoter was unmethylated in both cell subsets (Figure 1). However, the adjacent CpG shore was differentially methylated, with higher methylation levels in SSEA1+ cells. The analysis of data sets from various cancer types also showed a positive correlation between the methylation of the CpG shore with H1F0 mRNA levels. This CpG shore also appears to be an enhancer element, as it is enriched in H3K27ac and showed transactivating potential in reporter assays. Moreover, deletion of this region by CRISPR/CAS9 genome editing resulted in a decrease in the H1F0 mRNA levels. These results led the authors to conclude that H1.0 gene silencing in self-renewing tumor cells is, at least in part, due to the methylation of this enhancer region.
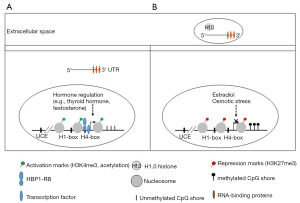
In addition to DNA methylation, other factors have been described to modulate H1.0 expression at transcriptional and post-transcriptional levels (Figure 1). At least three cis-acting regulatory elements, upstream control element (UCE), H1-box, and H4-box have been described in H1.0 gene promoter (15). HBP1, a high-mobility group box transcription factor, interacted specifically with the H4-box in H1F0 promoter. This transcription factor is ubiquitously expressed in a differentiation-dependent manner and interacts with the retinoblastoma protein (RB). Both HBP1 and RB were involved in the activation of H1.0 gene expression, providing a link between the cell cycle control machinery and cell differentiation signals. In human ESCs, the H1.0 promoter has bivalent chromatin marks (H3K27Me3 and H3K4Me3), suggesting that Polycomb-mediated repression may keep the H1.0 gene silent in stem cells (13). Core histone acetylation induced by treatment with histone deacetylase inhibitors such as trichostatin and butyrate has been shown to activate H1.0 gene expression during early Xenopus embryonic development (16).
Expression of H1.0 appears to be modulated by tissue-specific factors. In dendritic cells, which play an important role in antitumor immune response, the transcription factor NF-kappaB is actively involved in the regulation of H1.0 during differentiation (17). It has been shown that H1.0 expression in thyroid, adrenal cortex, and testes of rats is under hormone regulation (4). In rat brain, the expression of H1.0 in arcuate neurons is negatively regulated by the estradiol levels during the estrous cycle (18). Osmotic stimulation of supraoptic nucleus (SON) neurons of the rat hypothalamus induced a metabolic shift characterized by the overexpression of vasopressin, oxytocin and rRNA genes and by the depletion of H1.0 in chromatin (19). It thus appears that H1.0 is the only H1 subtype that can be controlled by external signals.
Post-transcriptional regulation of H1.0 in rat developing brain has also been described (20). H1.0 mRNA has been shown to interact by its 3’UTR with several RNA-binding proteins including heterogeneous nuclear ribonucleoprotein (hnRNPK and hnRNPA1), molecular chaperones (heat shock cognate 70, Hsc70), the cold shock domain-containing protein 2 (CSD-C2) and MYEF2 (21,22). H1.0 has been also been found in extracellular vessels (EVs) produced by oligodendrioma and melanoma cells, suggesting that cancer cells can escape differentiation by discarding this protein into EVs (22,23) (Figure 1). In addition in melanoma cells, EVs also contained H1.0 mRNA associated with RNA-binding proteins (22).
In summary, the results of Torres and colleagues together with evidences of numerous studies indicate a subtype-specific and important role of H1.0 in differentiation, as well as in cancer development. In the light of these findings, future studies are needed to completely understand H1.0 gene regulation in order to device therapeutic strategies for H1.0 re-expression in tumors. The results of Torres et al. are an important step forward towards the understanding of the relationships of chromatin epigenetics and tumor biology.
Acknowledgments
Funding: This work was supported by AGAUR (2014 SGR 1237), Spain.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Chen Qian (Center for Inflammation & Epigenetics, Houston Methodist Hospital Research Institute, Houston, TX, USA).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.03.06). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Torres CM, Biran A, Burney MJ, et al. The linker histone H1.0 generates epigenetic and functional intratumor heterogeneity. Science 2016;353. pii: aaf1644.
- Talbert PB, Ahmad K, Almouzni G, et al. A unified phylogeny-based nomenclature for histone variants. Epigenetics Chromatin 2012;21:5:7.
- Millán-Ariño L, Izquierdo-Bouldstridge A, Jordan A. Specificities and genomic distribution of somatic mammalian histone H1 subtypes. Biochim Biophys Acta 2016;1859:510-9.
- Gjerset R, Gorka C, Hasthorpe S, et al. Developmental and hormonal regulation of protein H1 degrees in rodents. Proc Natl Acad Sci U S A 1982;79:2333-7. [Crossref] [PubMed]
- Piña B, Martínez P, Simón L, et al. Differential kinetics of histone H1(0) accumulation in neuronal and glial cells from rat cerebral cortex during postnatal development. Biochem Biophys Res Commun 1984;123:697-702. [Crossref] [PubMed]
- Lea MA. Relationship of H1(0) histone to differentiation and cancer. Cancer Biochem Biophys 1987;9:199-209. [PubMed]
- Kostova NN, Srebreva LN, Milev AD, et al. Immunohistochemical demonstration of histone H1(0) in human breast carcinoma. Histochem Cell Biol. 2005;124:435-43. [Crossref] [PubMed]
- Warneboldt J, Haller F, Horstmann O, Danner BC, Füzesi L, Doenecke D, Happel N. Histone H1x is highly expressed in human neuroendocrine cells and tumours. BMC Cancer 2008;8:388. [Crossref] [PubMed]
- Jung Y, Joo KM, Seong DH, et al. Identification of prognostic biomarkers for glioblastomas using protein expression profiling. Int J Oncol 2012;40:1122-32. [PubMed]
- Scaffidi P. Histone H1 alterations in cancer. Biochim Biophys Acta 2016;1859:533-9.
- Medrzycki M, Zhang Y, McDonald JF, et al. Profiling of linker histone variants in ovarian cancer. Front Biosci (Landmark Ed) 2012;17:396-406. [Crossref] [PubMed]
- Cao K, Lailler N, Zhang Y, et al. High-resolution mapping of h1 linker histone variants in embryonic stem cells. PLoS Genet 2013;9:e1003417 [Crossref] [PubMed]
- Terme JM, Sesé B, Millán-Ariño L, et al. Histone H1 variants are differentially expressed and incorporated into chromatin during differentiation and reprogramming to pluripotency. J Biol Chem 2011;286:35347-57. [Crossref] [PubMed]
- Mayor R, Izquierdo-Bouldstridge A, Millán-Ariño L, et al. Genome distribution of replication-independent histone H1 variants shows H1.0 associated with nucleolar domains and H1X associated with RNA polymerase II-enriched regions. J Biol Chem 2015;290:7474-91. [Crossref] [PubMed]
- Lemercier C, Duncliffe K, Boibessot I, et al. Involvement of retinoblastoma protein and HBP1 in histone H1(0) gene expression. Mol Cell Biol 2000;20:6627-37. [Crossref] [PubMed]
- Seigneurin D, Grunwald D, Lawrence JJ, et al. Developmentally regulated chromatin acetylation and histone H1(0) accumulation. Int J Dev Biol 1995;39:597-603. [PubMed]
- Gabrilovich DI, Cheng P, Fan Y, et al. H1(0) histone and differentiation of dendritic cells. A molecular target for tumor-derived factors. J Leukoc Biol 2002;72:285-96. [PubMed]
- García-Segura LM, Luquín S, Martínez P, et al. Differential expression and gonadal hormone regulation of histone H1(0) in the developing and adult rat brain. Brain Res Dev Brain Res 1993;73:63-70. [Crossref] [PubMed]
- Lafarga M, García-Segura LM, Rodriguez JR, et al. Expression of histone H1 (zero) in transcriptionally activated supraoptic neurons. Brain Res Mol Brain Res 1995;29:317-24. [Crossref] [PubMed]
- Scaturro M, Cestelli A, Castiglia D, et al. Posttranscriptional regulation of H1 zero and H3.3B histone genes in differentiating rat cortical neurons. Neurochem Res 1995;20:969-76. [Crossref] [PubMed]
- Di Liegro CM, Schiera G, Proia P, et al. Identification in the rat brain of a set of nuclear proteins interacting with H1° mRNA. Neuroscience 2013;229:71-6. [Crossref] [PubMed]
- Schiera G, Di Liegro CM, Puleo V, et al. Extracellular vesicles shed by melanoma cells contain a modified form of H1.0 linker histone and H1.0 mRNA-binding proteins. Int J Oncol 2016;49:1807-14. [PubMed]
- Schiera G, Di Liegro CM, Saladino P, et al. Oligodendroglioma cells synthesize the differentiation-specific linker histone H1˚ and release it into the extracellular environment through shed vesicles. Int J Oncol 2013;43:1771-6. [PubMed]


