
Proteomics approaches to investigate cancer radiotherapy outcome: slow train coming
Introduction
Cancer is a one of the leading causes of mortality worldwide. Radiotherapy along with chemotherapy and surgery is considered as an essential therapeutic strategy against most tumour diseases. Although almost 60% of cancer patients are treated with radiotherapy (1) the effectiveness is not always guaranteed. Tumour radioresistance is the main reason for radiotherapy failure in cancer patients resulting in recurrence and metastasis. Therefore, a better understanding of the distinct molecular mechanisms behind radiosensitivity and radioresistance is urgently required to improve the outcome of cancer therapy.
A variety of established cutting-edge technologies applied in proteomics provide information on the molecular mechanisms that regulate cellular physiology and pathophysiology. In radiobiology studies, a broad range of quantitative proteomics platforms including chemical and metabolic labelling as well as label-free analysis have been used recently on different biomaterials ranging from organelles to cells and whole organs (2,3).
A comprehensive oncoproteomics analysis provides strong means to identify biological pathways involved in radiation-induced tissue toxicity and to quantify potential prognostic biomarkers for radioresistance. Advances in proteomics techniques improve the quality and accuracy of cancer treatment outcome requested in personalised medicine.
Proteomics approaches to identify predictive biomarkers of tumour radioresistance
The application of proteomics to investigate radiation response of different tumours has been a hot topic since more than a decade (reviewed by Lacombe et al. and Scaife et al.) (1,4,5), highlighting the importance of identifying predictive biomarkers of tumour radioresistance.
Feng et al. established a radioresistant subclone of nasopharyngeal carcinoma (NPC) cell line CNE2 by irradiating the cells with five subsequent sub-lethal doses (11 Gy, single dose). The proteome of surviving radioresistant cells in comparison to the proteome of the original cell line was analysed by two-dimensional electrophoresis (2DE) and mass spectrometry. The data showed differences in the expression of four proteins: 14-3-3σ, maspin, glucose-regulated protein (GRP-78), and Mn superoxide dismutase (MnSOD). These proteins were further validated in radiosensitive and radioresistant NPC tissues from patients (6).
In a similar study using radioresistant NPC cell line CNE1 that was generated after exposure to a single dose of 13 Gy, Zhang et al. identified 13 differentially expressed proteins resolved on 2DE. Among them, upregulation of heat shock protein 27 (HSP27) as a radioresistant biomarker was further validated using a panel of experiments including antisense oligonucleotides (ASOs), clonogenic survival assay, Hoechst 33258 staining of apoptotic cells, and MTT assay of cell viability. The authors showed that the decreased clonogenic survival and cell viability was associated with the inhibition of HSP27 expression by ASOs (7).
Wu et al. analysed radioresistant and radiosensitive NPC cancers using tissue biopsies. Applying 2DE proteomics, the authors found endoplasmic reticulum protein 29 (ERp29), Mn-SOD, HSP27 and glutathione-S-transferase (GST) ω1 significantly upregulated in radioresistant tissues. The association of ERp29 was further validated using immunohistochemistry analysis and knockdown strategy (8).
Chen et al. recently analysed the secretomes from radioresistant NPC cell line CNE-2R and radiosensitive parental cell line CNE-2 by isobaric tags for relative and absolute quantitation (iTRAQ) approach. Comparative proteomics between the two secretomes indicated 40 proteins significantly differentially expressed. Four differentially expressed secretory proteins (fibrillin-2, CD166, sulfhydryl oxidase 1 and cofilin-2) were verified by immunoblotting. These proteins could be considered as potential biomarkers for predicting NPC response to radiotherapy (9).
Using 2DE, Lin et al. compared the proteome of parental and radioresistant head and neck cancer (HNC) cell line created by fractionated radiation exposure (total dose of 60 Gy). Among 64 identified proteins potentially associated with radioresistance, Gp96, Grp78, HSP60, Rab40B, and GDF-15 were found upregulated and annexin V downregulated in the radioresistant cell line. The authors confirmed the role of Gp96 by showing that a Gp96 knockdown increased radiosensitivity in vitro and in vivo (10). Using similar experimental setting, Li et al. identified 16 differentially expressed proteins, among which Nm23 H1 was significantly upregulated whereas annexin A3 was significantly downregulated in resistant cells compared to control cells (11).
Skvortsov et al. used two-dimensional difference gel electrophoresis (2D-DIGE) to analyse the proteome of radioresistant FaDu-IRR and SCC25-IRR derived from two parental human head and neck squamous cell carcinoma cell lines after repeated exposure to ionizing radiation (10 Gy 10 times every 2 weeks). The majority of significantly deregulated proteins in both in FaDu-IRR and SCC25-IRR cells, if compared to the proteome of parental cells, belonged to Rac1 signalling pathways. The study showed that inhibition of Rac1 reduced the migration of these cells, thereby decreasing the capability to metastasise. Authors suggested Rac1 as a putative biomarker for head and neck squamous cell carcinoma radioresistance (12).
Using radioresistant oral squamous cell carcinoma (OSCC) cell lines induced by fractionated radiation with a total dose of 60 Gy, Lee et al. identified 18 cancer-related proteins changed in radioresistant cell lines. Among these proteins, the overexpression of NM23-H1 was further validated by immunoblotting and cDNA array. The authors suggested NM23-H1 as a biomarker to predict radioresistance in OSCC (13).
Kim et al. generated radioresistant laryngeal cancer cell lines from human HEp-2 cells using fractionated radiation (60 Gy/twice a week for 2 weeks). The proteomics analysis showed 16 proteins significantly changed between HEp-2 and RR-HEp-2 cells. The expression of chloride intracellular channel 1 (CLIC1) that was markedly downregulated in RR-HEp-2 cells was further validated by immunoblotting and immunohistochemistry. Inhibition of CLIC1 in HEp-2 cells resulted in increased radioresistance by suppressing the radiation-induced cellular reactive oxygen species (ROS) levels (14).
Using multiple-reaction monitoring (MRM) analysis, Guo et al. studied the global kinome of radioresistant human breast cancer cell line MCF-7/C6 and its parental MCF-7 cell line. The analysis showed a significant deregulation of several kinases involved in cell cycle progression and DNA damage response, suggesting that kinases in general play an important role in tumour radioresistance (15).
Kim et al. used stable isotope labelling by amino acids in cell culture (SILAC) method to investigate the proteome of human breast cancer cell line (MDA-MB-231) 48 h after single or fractionated radiation (total dose of 10 Gy). The interaction analysis of highest ranking upregulated proteins (cathepsin D, gelsolin, argininosuccinate synthase 1, peroxiredoxin 5, and C-type mannose receptor 2) showed that these proteins are associated with cell adhesion and metastasis in tumour cells (16).
Yun et al. recently established a radioresistant lung cancer cell line from parental radiosensitive cells by fractionated exposure (2 Gy twice a week for 20 weeks). The proteomics comparison of these two cell lines using 2DE showed significant upregulation of four candidate proteins functioning as putative radioresistance biomarkers: plasminogen activator inhibitor 2 (PAI-2), nodal modulator 2 (NOMO2), kinesin light chain 4 (KLC4), and procollagen-lysine 2-oxoglutarate 5-dioxygenase 3 (PLOD3) (17).
Since a large amount of non-small cell lung cancer (NSCLC) patients are treated by irradiation after chemotherapy failure Wei et al. investigated whether multidrug resistance possibly increases radioresistance. Applying 2DE, the authors compared lung cancer cell line A549 and cisplatin-resistant A549 cell line after exposure to 6 Gy. Among radiation-induced significantly deregulated proteins in drug resistant cells 4 up-regulated proteins (HSPB1, vimentin, cofilin-1, and annexin A4) were verified by immunoblotting. The authors further confirmed the increased expression level of these proteins in formalin-fixed, paraffin-embedded (FFPE) tissue by immunohistochemistry (18).
Using label-free quantification method, Hao et al. compared the proteome of radioresistant and radioresponsive prostate cancer (PC-3) xenografts raised in a mouse model after exposure to 2 Gy/day for five consecutive days. The pathway analysis indicated that glycolysis was the most important pathway deregulated in the radioresistant tissue. The authors showed that inhibition of the glycolysis marker lactate dehydrogenase A (LDHA) sensitised PC-3RR cells to radiotherapy (19).
The potential biomarkers identified in radioresistance tumours by proteomics are summarised in Table 1. The obvious lack of universal biomarkers between different cancer types reflects the complexity and heterogeneity of tumour response to irradiation. Although biomarkers found in various studies show no consensus, they belong to common protein families. Chaperones (HSP27, HSP60, HSPB1 and GRP78), antioxidant proteins (Mn-SOD and peroxiredoxin) and structural proteins (annexin, vimentin and gelsolin) were often identified as differentially regulated in radioresistant cell lines in different studies. The altered proteins were involved in a variety of cellular events including cell cycle, signal transduction and stress response. As shown in Table 1, the majority of the proteomics studies presented here were searching for radioresistance biomarkers of head and neck cancer, probably due to availability and suitability of the biomaterial.
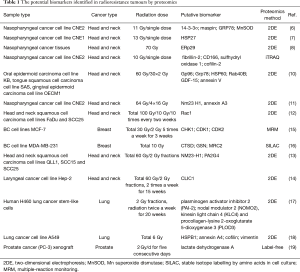
Full table
Proteomic approaches to study radiation-induced normal tissue toxicity
The main challenge in cancer radiotherapy is to choose the radiation dose effective enough to kill the tumour but to minimise damage to the surrounding normal tissue. Radiation-induced late toxicity limits the frequency and thereby the efficiency of the treatment (20). Normal cell killing, increased fibrosis, excessive ROS production leading to oxidation of lipids and proteins, massive inflammation due to enhanced production of pro-inflammatory cytokines, and misbalanced cellular metabolism are all adverse side effects of irradiation contributing to normal tissue toxicity (2,21). Application of proteomics facilitates the identification of molecular mechanisms involved in normal tissue injury and thereby provides means to preventive measures.
Radiation-induced alterations in the cellular proteome (in vitro proteomics)
Different human cell lines have been used as models to investigate radiation-induced proteome alterations in normal tissue. Sriharshan and colleagues studied the immediate radiation-induced alterations in the proteome of human endothelial cell line (EA.hy926) imitating rapid radiation-induced vascular damage. The effect of an acute gamma dose of 2.5 Gy was investigated 4 and 24 h post-radiation (22). Using two complementary proteomics approaches, SILAC and 2D-DIGE, significant changes in several biological pathways such as glycolysis, oxidative phosphorylation, Rho-mediated cell motility and non-homologous end joining were identified. The study showed that alteration in proteins involved in metabolic activity, stress response and apoptosis were immediate (Jeny4 h). Cellular signalling and transcriptional activity were the most affected pathways at 24 h (22).
In a study investigating mechanisms behind individual radiation sensitivity Skiöld et al. compared proteomes of leukocytes isolated from in vitro irradiated whole blood from normosensitive or extremely sensitive patients. The leukocyte proteomes from non-irradiated blood samples from the same patients were used as controls (23). This analysis showed markedly altered expression of proteins involved in the oxidative stress response, coagulation and acute phase response in the sensitive patients. This study suggested that the impairment in the redox balance and differences in the inflammatory response regulated by the transcription factor peroxisome proliferator-activated receptor alpha (PPAR alpha) hallmark individual radiation sensitivity (23).
Radiation-induced alterations in the tissue proteome (in vivo proteomics)
Data obtained from cellular experiments alone is not sufficient in order to understand complex and systemic tissue responses. In spite of several technical challenges, tissue proteomics has recently made great advances in radiobiology.
Ionizing radiation is a well-documented risk factor of heart disease in the treatment of cancers of the thorax area as reviewed recently (24). Understanding the mechanism of radiation-induced heart failure and cardiac vascular impairment is essential to improve therapeutic strategies in order to avoid heart injury. To investigate the short term (Jeny4 and Jeny24 h) effect of total body irradiation of 3 Gy (gamma) on the cardiac proteome of C57Bl/6 mice, Azimzadeh et al. used two complementary quantitative proteomics approaches: isotope-coded protein label (ICPL) and minimal 2D-DIGE (25). Proteome profiling showed that this radiation dose immediately induced biological responses in the heart including inflammation, oxidative response, and remodelling of structural proteins. The analysis indicated mitochondrial proteins as the most radiation sensitive protein category in the heart (25).
Barjaktarovic et al. used ICPL and saturated 2D-DIGE to analyse the cardiac mitochondrial proteome of C57Bl/6 mice 4 weeks after local heart exposure to X-rays (0.2 or 2 Gy) (26). Proteome profiling indicated that alterations in oxidative phosphorylation, pyruvate metabolism, and cytoskeletal structure were stable several weeks after the exposure to low- or high-dose ionizing radiation (26). The radiation effect in cardiac mitochondria was also analysed 40 weeks after local heart exposure using similar X-ray doses (0.2 or 2 Gy) (27). The proteome analysis showed that the number of differentially regulated proteins as well as the magnitude of protein expression alteration was increased if compared to the data at 4 weeks. These findings suggested that especially the impairment of oxidative phosphorylation is progressive and contributed to the long-term impairment of cardiac energy production (27).
Bakshi et al. studied the alteration of murine cardiac and hepatic proteomes after exposure to low and moderate radiation doses (0.02, 0.1, 0.5, and 1.0 Gy) (28,29). In these studies, NMRI mice received single doses of total body 60Co gamma-radiation on postnatal day 10 (PND10) and were sacrificed 7 months later. Functional analysis of the cardiac proteome showed that most of the deregulated proteins were involved in metabolic processes, inflammatory response, and cytoskeletal structure (29). The study on hepatic proteome of the same mice showed that these radiation doses caused immediate (Jeny24 h) inhibition of the glycolysis pathway and pyruvate dehydrogenase availability in the liver that in turn resulted in significant long-term alterations in hepatic lipid metabolism and increased inflammation (28). The radiation-induced change in both heart and liver proteome was associated with altered activation of the transcription factor PPAR alpha, the major regulator of lipid metabolism (28,29).
This was in agreement with previous data similarly showing alteration in PPAR alpha-regulated proteins in murine cardiac tissue 16 weeks after local high-dose radiation (8 and 16 Gy X-ray) (29). This study using C57Bl/6 mice showed significant expression changes in proteins involved in lipid metabolism and energy production. In addition, marked changes were shown in the expression of the PPAR alpha target genes most of which are essential for orderly functioning of energy metabolism and mitochondrial respiratory chain. The study emphasized the role of PPAR alpha as a novel target of ionizing radiation. The impairment of this transcriptional regulator contributes to the heart pathology after radiation exposure (29).
Recently, Subramanian et al. showed persistent alteration of the cardiac proteome and transcriptome in C57Bl/6 mice 40 weeks after local high-dose heart exposure (16 Gy X-ray). The integrated network analysis of transcriptomics and proteomics data showed that transforming growth factor (TGF) beta signalling and PPAR alpha signalling were affected by irradiation both at gene and protein level. The data indicated induction of TGF beta signalling but inactivation of PPAR alpha signalling in the irradiated heart (30).
Azimzadeh et al. analysed cardiac endothelial cells harvested from mice treated with local heart high-dose radiation (8 and 16 Gy X-ray) (31). The proteomics analysis using ICPL indicated radiation-induced endothelial dysfunction in the irradiated cells that was characterised by impaired energy metabolism and perturbation of the insulin/IGF-PI3K-Akt signalling pathway. These data also provided evidence for premature endothelial senescence, increased oxidative stress, decreased NO availability and enhanced inflammation as main causes of radiation-induced long-term vascular dysfunction (31).
High doses of ionizing radiation as used in the context of brain cancer therapy are known to have adverse effects on the brain as recently reviewed (32). The effect of low radiation doses is less clear but a possible connection between ionizing radiation and neurodegenerative diseases has been suggested (33).
Kempf et al. studied the effect of acute gamma doses of 0.5, 1.0 and 4.0 Gy on mouse hippocampal neuronal HT22 cells immediately after the exposure (Jeny4 and Jeny24 h) (34). To compare the findings with the in vivo response, male NMRI mice were irradiated on the PND10 with a gamma dose of 1.0 Gy and the proteome alteration in hippocampus and cortex was analysed 24 h post-irradiation. The cellular proteome study showed significant changes in the signalling pathways related to synaptic actin remodelling at 1.0 and 4.0 Gy but not at 0.5 Gy. Alterations in similar pathways were observed in the irradiated hippocampus and cortex (34).
In a similar study, the immediate (Jeny24 h) proteome response of hippocampus and cortex was analysed in female C57BL/6J mice irradiated on the PND10 with gamma doses of 0.1 or 0.5 Gy (35). The analysis showed that the levels of proteins involved in the mitochondrial and synaptic functions were rapidly changed. The data suggested that this alteration adversely affected the mitochondrial function, number of dendritic spines and neurite outgrowth predominately in the cortex already at 0.1 Gy but also in the hippocampus at 0.5 Gy (35).
Using ICPL approach, the long-term effects of moderate doses (0.5 or 1.0 Gy) of total body gamma radiation were studied on the cortical and hippocampal proteome of neonatally exposed NMRI mice 6–7 months after the exposure. The long-term proteomics analysis confirmed previously seen immediate alteration of the signalling pathways related to synaptic actin remodelling. The radiation-induced alteration in the Rac1-Cofilin pathway essential for the formation of synapses was suggested to be associated with the cognitive impairment observed in these animals (36). The hippocampal and cortical downregulation of Rac1, upregulation of cofilin and upregulation of phosphorylated cAMP response element-binding protein (CREB) was found in both of these low-dose long-term studies independent of the mouse strain or gender (36,37). The changes in the Rac1-cofilin and CREB pathways could be considered as a radiation-induced proteomic fingerprint of neonatal irradiation in the mouse brain.
To investigate the role of the age at exposure in the brain function, the proteome alterations were measured using ICPL in the hippocampus of C57Bl/6 mice irradiated cranially either at PND10 or week 10 using low or moderate doses (0.1 and 2 Gy). The study clearly indicated an increased sensitivity to cranial irradiation at the early age compared to the young adult period. Still, the analysis indicated significant common changes in the hippocampus at both irradiation time points including alteration in oxidative phosphorylation, mitochondrial dysfunction and superoxide radical degradation (38).
Hengel et al. recently studied the proteome response of reconstituted human skin tissue model 8 h after treatment with 0.1 Gy (X-ray) using 8-plex iTRAQ labelling. Functional analysis of proteomics data indicated significant changes in the biological pathways involved in the cellular structure and actin cytoskeleton (39). This study confirmed the previous observation about the effect of low-dose irradiation on the expression and post-translational modification (PTM) of the skin barrier protein, filaggrin (40).
Radiation-induced alterations in protein levels of bio fluids identified by proteomics
Bio fluids are reflecting the entire tissue ensemble present in a patient, providing information on the pathophysiology of a specific disease or disease state (41). In spite of the complexity of serum and urine proteome, most of the biomarker studies using proteomics have used bio fluids due to the relatively easy and non-invasive manner to collect samples. Radiation-induced proteomic alterations in different bio fluids have been recently reviewed (42,43).
Using a bioinformatics approach, Oh et al. identified serum biomarkers of lung injury during and after radiotherapy. The authors analysed the serum from 26 locally advanced NSCLC patients treated with radiotherapy compared to age, gender, and ethnicity matched control cohort. A graph-based computational method suggested increased level of macroglobulin alpha 1 as a predictive biomarker for radiation-induced pneumonitis (44).
2D-DIGE and surface-enhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI-TOF-MS) techniques were applied to analyse serum proteins after a high-dose (20, 40 and 80 Gy gamma) localised irradiation of the skin (43). The authors found an upregulation of 15 proteins involved in glycosylation in the liver and increased serum cytokines suggesting a systemic response to radiation exposure (45).
Using 2D-DIGE, Nylund et al. profiled the proteome changes in the plasma of 30 locally exposed clinical patients that had received fractionated radiation therapy. The results were compared to those received using plasma samples from three radiological accident victims exposed in 1994. The study showed no differences in the protein expression between pre- and post-radiation exposure in the clinical cases but the expression of haptoglobin and serotransferrin/transferrin was downregulated in the plasma from radiological accident victims (46).
Huang et al. used 2DE to analyse sera from 37 NSCLC patients who were divided into sensitive and resistant groups based on the radiotherapy outcome (doses ranging from 62.4 to 68.0 Gy) and further treated with cisplatin-based chemotherapy. Among eight differently expressed proteins, six proteins were successfully identified. Five of these were higher expressed and one lower expressed in resistant group in comparison to the sensitive group. Among these potential radioresistance biomarkers, the increased level of alpha-1-antitrypsin (α1-AT) was successfully validated in additional 50 serum samples (47).
To investigate the biological processes involved in response to partial body irradiation during cancer treatment, Widlak et al. analysed the serum proteome of 20 head and neck squamous cell cancer patients that had received radiotherapy. The significantly upregulated proteins were associated with acute phase response and inflammation (48).
To identify biomarkers of radiation-induced lung toxicity, Cai et al. analysed the plasma proteome of NSCLC patients with and without radiation-induced lung toxicity (RILT2). The patients received radiotherapy within minimum follow-up of 1 year. Median radiation doses were 66 and 67.5 Gy for patients with and without RILT2, respectively. The analysis indicated that C4b-binding protein alpha chain and vitronectin were significantly upregulated whereas the immunoglobulin (Ig) kappa chain V-III region Ti and region HAH were significantly downregulated in patients with RILT2 compared to patients without RILT2. The upregulated proteins belonged to the inflammatory networks of TGF-beta-1 and interleukin-8 that are known to play an important role in the induction of radiation-induced lung damage (49).
Proteomics studies using formalin-fixed paraffin-embedded (FFPE) samples
Clinical tissue banks represent an invaluable source of biological information, especially if information on diagnosis and outcome is available (50). Samples from clinical archives can be used for retrospective validation of biomarkers of radiation exposure, prognosis and disease. For this reason, proteomics analysis of FFPE tissue has recently gained increased attention in the proteomics society (51,52). However, the conventional protein labelling often used in proteomics is a suboptimal method for protein quantification in the case of FFPE tissue, mainly due to difficulties generated by fixation process leading to protein cross-linking (53). Protocols to study gene, miRNA or protein expression using FFPE samples have been developed in recent years, especially in the field of radiobiology (54-56).
Sepiashvili et al. compared the proteome profiles of human papillomavirus (HPV)-positive (n=27) and HPV-negative (n=26) FFPE biopsies from oropharyngeal carcinoma patients. The analysis showed 174 differently expressed proteins between the two groups. The deregulated proteins were involved in the regulation of cell cycle, DNA replication, apoptosis, and immune response. Using big scale integration analysis, the authors found increased level of the oncoprotein cortactin in HPV-negative biopsies. Cortactin is a potential biomarker for radioresistance in HPV-negative samples and may contribute to reduced survival in the patients (57).
Outlook and future perspectives
Considering the accruing data from different oncoproteomics studies over the last few years, the verification of potential biomarkers is a necessity. Only a few of the reported biomarker proteins have passed into the clinical validation phase. The time has come to carefully explore the clinical relevance of at least the most promising protein biomarkers.
On the other hand, many proteomics studies, especially those performed to identify biomarkers of radiotherapy resistance, do not represent state-of-the-art proteomics approaches. The current proteomics platforms from global high throughput screening to targeted mass spectrometry using MRM or selective reaction monitoring (SRM) provide highly specific and sensitive tools for proteome analysis. The recent developments in proteomics software and hardware overcome the complex of problems from old conventional proteomic analyses. Thus, the search for biomarkers in oncoproteomics has not yet reached the final phase and needs to be continued.
In this context, the radiation-dependent alterations in PTM of proteins have rarely been addressed in the radiotherapy biomarker studies. As both cancer and normal tissue response to irradiation are regulated by PTM, identification and examination of most common modifications such as phosphorylation, ubiquitination, lysine acetylation and methylation in clinical samples will certainly provide a better understanding for cellular response and resistance to radiation exposure.
The broad and diverse range of protein classes reported here suggests that the application of multiple proteomic biomarkers rather than a single protein might prove to be more accurate in the prognosis of radiotherapy outcome. As high-throughput proteomic analysis is already now technically feasible it is to be expected that the slow start in the usage of this powerful technique in the field of oncoproteomics will turn into a frequently used tool in the clinics in the near future.
Acknowledgments
The authors thank Bob Dylan for the album “Slow Train Coming” that inspired to create the title of this review.
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Marco Durante, Giusi I. Forte, Giorgio Russo) for the series “Radiobiological models towards a personalized radiation oncology” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.03.83). The series “Radiobiological models towards a personalized radiation oncology” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lacombe J, Azria D, Mange A, et al. Proteomic approaches to identify biomarkers predictive of radiotherapy outcomes. Expert Rev Proteomics 2013;10:33-42. [Crossref] [PubMed]
- Azimzadeh O, Atkinson MJ, Tapio S. Proteomics in radiation research: present status and future perspectives. Radiat Environ Biophys 2014;53:31-8. [Crossref] [PubMed]
- Chang L, Graham P, Hao J, et al. Proteomics discovery of radioresistant cancer biomarkers for radiotherapy. Cancer Lett 2015;369:289-97. [Crossref] [PubMed]
- Lacombe J, Mange A, Azria D, et al. Identification of predictive biomarkers to radiotherapy outcome through proteomics approaches. Cancer Radiother 2013;17:62-9; quiz 70, 72.
- Scaife L, Hodgkinson VC, Drew PJ, et al. Differential proteomics in the search for biomarkers of radiotherapy resistance. Expert Rev Proteomics 2011;8:535-52. [Crossref] [PubMed]
- Feng XP, Yi H, Li MY, et al. Identification of biomarkers for predicting nasopharyngeal carcinoma response to radiotherapy by proteomics. Cancer Res 2010;70:3450-62. [Crossref] [PubMed]
- Zhang B, Qu JQ, Xiao L, et al. Identification of heat shock protein 27 as a radioresistance-related protein in nasopharyngeal carcinoma cells. J Cancer Res Clin Oncol 2012;138:2117-25. [Crossref] [PubMed]
- Wu P, Zhang H, Qi L, et al. Identification of ERp29 as a biomarker for predicting nasopharyngeal carcinoma response to radiotherapy. Oncol Rep 2012;27:987-94. [PubMed]
- Chen ZT, Li L, Guo Y, et al. Analysis of the differential secretome of nasopharyngeal carcinoma cell lines CNE-2R and CNE-2. Oncol Rep 2015;34:2477-88. [PubMed]
- Lin TY, Chang JT, Wang HM, et al. Proteomics of the radioresistant phenotype in head-and-neck cancer: Gp96 as a novel prediction marker and sensitizing target for radiotherapy. Int J Radiat Oncol Biol Phys 2010;78:246-56. [Crossref] [PubMed]
- Li L, Huang S, Zhu X, et al. Identification of radioresistance-associated proteins in human nasopharyngeal carcinoma cell lines by proteomic analysis. Cancer Biother Radiopharm 2013;28:380-4. [Crossref] [PubMed]
- Skvortsov S, Jimenez CR, et al. Radioresistant head and neck squamous cell carcinoma cells: intracellular signaling, putative biomarkers for tumor recurrences and possible therapeutic targets. Radiother Oncol 2011;101:177-82. [Crossref] [PubMed]
- Lee SY, Park HR, Cho NH, et al. Identifying genes related to radiation resistance in oral squamous cell carcinoma cell lines. Int J Oral Maxillofac Surg 2013;42:169-76. [Crossref] [PubMed]
- Kim JS, Chang JW, Yun HS, et al. Chloride intracellular channel 1 identified using proteomic analysis plays an important role in the radiosensitivity of HEp-2 cells via reactive oxygen species production. Proteomics 2010;10:2589-604. [Crossref] [PubMed]
- Guo L, Xiao Y, Fan M, et al. Profiling global kinome signatures of the radioresistant MCF-7/C6 breast cancer cells using MRM-based targeted proteomics. J Proteome Res 2015;14:193-201. [Crossref] [PubMed]
- Kim MH, Jung SY, Ahn J, et al. Quantitative proteomic analysis of single or fractionated radiation-induced proteins in human breast cancer MDA-MB-231 cells. Cell Biosci 2015;5:2. [Crossref] [PubMed]
- Yun HS, Baek JH, Yim JH, et al. Radiotherapy diagnostic biomarkers in radioresistant human H460 lung cancer stem-like cells. Cancer Biol Ther 2016;17:208-18. [Crossref] [PubMed]
- Wei R, Zhang Y, Shen L, et al. Comparative proteomic and radiobiological analyses in human lung adenocarcinoma cells. Mol Cell Biochem 2012;359:151-9. [Crossref] [PubMed]
- Hao J, Graham P, Chang L, et al. Proteomic identification of the lactate dehydrogenase A in a radioresistant prostate cancer xenograft mouse model for improving radiotherapy. Oncotarget 2016;7:74269-85. [PubMed]
- Shukla HD, Mahmood J, Vujaskovic Z. Integrated proteo-genomic approach for early diagnosis and prognosis of cancer. Cancer Lett 2015;369:28-36. [Crossref] [PubMed]
- Kim JH, Jenrow KA, Brown SL. Mechanisms of radiation-induced normal tissue toxicity and implications for future clinical trials. Radiat Oncol J 2014;32:103-15. [Crossref] [PubMed]
- Sriharshan A, Boldt K, Sarioglu H, et al. Proteomic analysis by SILAC and 2D-DIGE reveals radiation-induced endothelial response: Four key pathways. J Proteomics 2012;75:2319-30. [Crossref] [PubMed]
- Skiöld S, Azimzadeh O, Merl-Pham J, et al. Unique proteomic signature for radiation sensitive patients; a comparative study between normo-sensitive and radiation sensitive breast cancer patients. Mutat Res 2015;776:128-35. [Crossref] [PubMed]
- Tapio S. Pathology and biology of radiation-induced cardiac disease. J Radiat Res 2016;57:439-48. [Crossref] [PubMed]
- Azimzadeh O, Scherthan H, Sarioglu H, et al. Rapid proteomic remodeling of cardiac tissue caused by total body ionizing radiation. Proteomics 2011;11:3299-311. [Crossref] [PubMed]
- Barjaktarovic Z, Schmaltz D, Shyla A, et al. Radiation-induced signaling results in mitochondrial impairment in mouse heart at 4 weeks after exposure to X-rays. PLoS One 2011;6:e27811 [Crossref] [PubMed]
- Barjaktarovic Z, Shyla A, Azimzadeh O, et al. Ionising radiation induces persistent alterations in the cardiac mitochondrial function of C57BL/6 mice 40 weeks after local heart exposure. Radiother Oncol 2013;106:404-10. [Crossref] [PubMed]
- Bakshi MV, Azimzadeh O, Barjaktarovic Z, et al. Total body exposure to low-dose ionizing radiation induces long-term alterations to the liver proteome of neonatally exposed mice. J Proteome Res 2015;14:366-73. [Crossref] [PubMed]
- Bakshi MV, Barjaktarovic Z, Azimzadeh O, et al. Long-term effects of acute low-dose ionizing radiation on the neonatal mouse heart: a proteomic study. Radiat Environ Biophys 2013;52:451-61. [Crossref] [PubMed]
- Subramanian V, Seemann I, Merl-Pham J, et al. The Role of TGF Beta and PPAR Alpha Signalling Pathways in Radiation Response of Locally Exposed Heart: Integrated Global Transcriptomics and Proteomics Analysis. J Proteome Res 2017;16:307-18. [Crossref] [PubMed]
- Azimzadeh O, Sievert W, Sarioglu H, et al. Integrative proteomics and targeted transcriptomics analyses in cardiac endothelial cells unravel mechanisms of long-term radiation-induced vascular dysfunction. J Proteome Res 2015;14:1203-19. [Crossref] [PubMed]
- Hladik D, Tapio S. Effects of ionizing radiation on the mammalian brain. Mutat Res 2016;770:219-30. [Crossref] [PubMed]
- Kempf SJ, Azimzadeh O, Atkinson MJ, et al. Long-term effects of ionising radiation on the brain: cause for concern? Radiat Environ Biophys 2013;52:5-16. [Crossref] [PubMed]
- Kempf SJ, Buratovic S, von Toerne C, et al. Ionising radiation immediately impairs synaptic plasticity-associated cytoskeletal signalling pathways in HT22 cells and in mouse brain: an in vitro/in vivo comparison study. PLoS One 2014;9:e110464 [Crossref] [PubMed]
- Kempf SJ, Moertl S, Sepe S, et al. Low-dose ionizing radiation rapidly affects mitochondrial and synaptic signaling pathways in murine hippocampus and cortex. J Proteome Res 2015;14:2055-64. [Crossref] [PubMed]
- Kempf SJ, Casciati A, Buratovic S, et al. The cognitive defects of neonatally irradiated mice are accompanied by changed synaptic plasticity, adult neurogenesis and neuroinflammation. Mol Neurodegener 2014;9:57. [Crossref] [PubMed]
- Kempf SJ, Sepe S, von Toerne C, et al. Neonatal Irradiation Leads to Persistent Proteome Alterations Involved in Synaptic Plasticity in the Mouse Hippocampus and Cortex. J Proteome Res 2015;14:4674-86. [Crossref] [PubMed]
- Casciati A, Dobos K, Antonelli F, et al. Age-related effects of X-ray irradiation on mouse hippocampus. Oncotarget 2016;7:28040-58. [PubMed]
- Hengel SM, Aldrich JT, Waters KM, et al. Quantitative Proteomic Profiling of Low-Dose Ionizing Radiation Effects in a Human Skin Model. Proteomes 2014;2:382-98. [Crossref] [PubMed]
- Yang F, Waters KM, Webb-Robertson BJ, et al. Quantitative phosphoproteomics identifies filaggrin and other targets of ionizing radiation in a human skin model. Exp Dermatol 2012;21:352-7. [Crossref] [PubMed]
- Veenstra TD, Conrads TP, Hood BL, et al. Biomarkers: mining the biofluid proteome. Mol Cell Proteomics 2005;4:409-18. [Crossref] [PubMed]
- Pernot E, Hall J, Baatout S, et al. Ionizing radiation biomarkers for potential use in epidemiological studies. Mutat Res 2012;751:258-86. [Crossref] [PubMed]
- Guipaud O. Serum and plasma proteomics and its possible use as detector and predictor of radiation diseases. Adv Exp Med Biol 2013;990:61-86. [Crossref] [PubMed]
- Oh JH, Craft JM, Townsend R, et al. A bioinformatics approach for biomarker identification in radiation-induced lung inflammation from limited proteomics data. J Proteome Res 2011;10:1406-15. [Crossref] [PubMed]
- Chaze T, Hornez L, Chambon C, et al. Serum Proteome Analysis for Profiling Predictive Protein Markers Associated with the Severity of Skin Lesions Induced by Ionizing Radiation. Proteomes 2013;1:40-69. [Crossref] [PubMed]
- Nylund R, Lemola E, Hartwig S, et al. Profiling of low molecular weight proteins in plasma from locally irradiated individuals. J Radiat Res 2014;55:674-82. [Crossref] [PubMed]
- Huang W, Ding X, Li B, et al. Serum biomarkers analyzed by LC-MS/MS as predictors for short outcome of non-small cell lung cancer patients treated with chemoradiotherapy. Neoplasma 2013;60:11-8. [Crossref] [PubMed]
- Widlak P, Jelonek K, Wojakowska A, et al. Serum Proteome Signature of Radiation Response: Upregulation of Inflammation-Related Factors and Downregulation of Apolipoproteins and Coagulation Factors in Cancer Patients Treated With Radiation Therapy--A Pilot Study. Int J Radiat Oncol Biol Phys 2015;92:1108-15. [Crossref] [PubMed]
- Cai XW, Shedden KA, Yuan SH, et al. Baseline plasma proteomic analysis to identify biomarkers that predict radiation-induced lung toxicity in patients receiving radiation for non-small cell lung cancer. J Thorac Oncol 2011;6:1073-8. [Crossref] [PubMed]
- Tapio S, Atkinson MJ. Molecular information obtained from radiobiological tissue archives: achievements of the past and visions of the future. Radiat Environ Biophys 2008;47:183-7. [Crossref] [PubMed]
- Gustafsson OJ, Arentz G, Hoffmann P. Proteomic developments in the analysis of formalin-fixed tissue. Biochim Biophys Acta 2015;1854:559-80. [Crossref] [PubMed]
- Magdeldin S, Yamamoto T. Toward deciphering proteomes of formalin-fixed paraffin-embedded (FFPE) tissues. Proteomics 2012;12:1045-58. [Crossref] [PubMed]
- Azimzadeh O, Barjaktarovic Z, Aubele M, et al. Formalin-fixed paraffin-embedded (FFPE) proteome analysis using gel-free and gel-based proteomics. J Proteome Res 2010;9:4710-20. [Crossref] [PubMed]
- Azimzadeh O, Scherthan H, Yentrapalli R, et al. Label-free protein profiling of formalin-fixed paraffin-embedded (FFPE) heart tissue reveals immediate mitochondrial impairment after ionising radiation. J Proteomics 2012;75:2384-95. [Crossref] [PubMed]
- Ludyga N, Grunwald B, Azimzadeh O, et al. Nucleic acids from long-term preserved FFPE tissues are suitable for downstream analyses. Virchows Arch 2012;460:131-40. [Crossref] [PubMed]
- Azimzadeh O, Azizova T, Merl-Pham J, et al. A dose-dependent perturbation in cardiac energy metabolism is linked to radiation-induced ischemic heart disease in Mayak nuclear workers. Oncotarget 2017;8:9067-78. [PubMed]
- Sepiashvili L, Waggott D, Hui A, et al. Integrated omic analysis of oropharyngeal carcinomas reveals human papillomavirus (HPV)-dependent regulation of the activator protein 1 (AP-1) pathway. Mol Cell Proteomics 2014;13:3572-84. [Crossref] [PubMed]


