
Peroxiredoxin-4 as a potential biomarker of early gastric cancer screened by Cell-SELEX
Introduction
Gastric cancer is currently the fifth most common cancer worldwide, accounting for 7% of all newly diagnosed cancer cases and 9% of all cancer-related deaths (1). Because more than half of all patients with gastric cancer are diagnosed at late stages of the disease, post-operative survival rates remain low (2). Therefore, early diagnosis is essential to identify and select the most appropriate treatment in order to reduce mortality. The development of gastric cancer is a multi-factor and multi-channel process; So far, some serum biomarkers have been identified to be useful for specifically targeting stomach cancer, such as CA19-9 and CEA. However, the sensitivity and specificity of these tumor markers is not very high (3). And some biomarkers may be useful only when it comes to the prognosis of gastric cancer (4). Some serologic testing methods, including serum pepsinogen and Helicobacter pylori antibody testing, are closely related to the diseases process and disease prognosis in patients with gastric cancer. However, these methods which are applied in the clinical stage show minimal effectiveness in diagnose of early gastric cancer (5,6). In this study, we aimed to find a new biomarker that can diagnose gastric cancer in the early stage.
Systematic evolution of ligands by exponential enrichment (SELEX) (7,8) is an in vitro screening technique utilizing large-capacity, random oligonucleotide libraries synthesized by combinatorial or traditional chemical methods. In each round of selection, we gathered those aptamers which can tightly bind to the target. And with the help of polymerase chain reaction (PCR), those aptamers would be amplified for the next round of selection. After several rounds, we can found the aptamers with high sensitivity and specificity (9). Aptamers are single-stranded oligonucleotides (10), which bind to the surfaces of various targets by folding in stable three-dimensional structures, and are therefore very useful in molecular recognition. Compared to antibodies, aptamers show similar or higher level affinity; furthermore, aptamers are considered to be better capturing agents for small molecules, because (I) their shorter size more accurately discriminates between functional groups with similar structures; and (II) aptamers targeting small molecules can be selected in vitro without requiring haptens, which are needed for selection of antibodies against molecules (11).
Recently, aptamers have been applied in diagnostic capacities for fundamental research experiments, disease diagnosis (11), clinical treatment (12,13), and drug screening (14). Cell-SELEX screening is a specific type of SELEX technology. Cell-SELEX screening uses the differences in the surface molecules between any two complete, living cell types to identify aptamers that are specific to target cells. In clinical practice, this allows for accurate identification and for distinguishing between normal and target cell lines. The objective of the present study was to use cell-SELEX technology to identify early gastric cancer biomarkers. This information should facilitate future studies aimed at understanding the mechanisms underlying the development of gastric cancer and differences in surface molecule expression in different tissues, for the purpose of improving early diagnosis.
Since the establishment of cell-SELEX screening, it has been used for laboratory diagnosis using a large number of oligonucleotide aptamer libraries (15). This approach utilizes single-stranded DNA (ssDNA) aptamers with high sensitivity, affinity, specificity, and additional advantages. To date, lymphocytic leukemia, myeloid leukemia, liver cancer, small-cell lung cancer, colorectal cancer, and other tumor cell-specific aptamers have been identified using the cell-SELEX approach (16,17).
In the present study, cell-SELEX technology was utilized to identify aptamers specific to early gastric cancer primary cells. Using streptavidin-biotin magnetic beads (18), a secondary library of highly specific ssDNA aptamers was prepared for the clinical detection of early gastric cancer and the isolation of cell-surface marker molecules. Target proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) mass spectrometry. We further selected the target protein with the highest affinity and specificity and determined its expression in the early stage of gastric cancer.
Methods
Extraction and identification of early gastric cancer cells
All participants provided informed written consent for inclusion in this study. The study was approved by the School of Basic Medical Ethics Committee, Lanzhou University. Patients who underwent radical operation for the treatment of early gastric cancer at the People’s Hospital of Wuwei City and the Department of General Surgery of the Second Hospital of Lanzhou University were selected for inclusion. A total of 210 patients were identified in this study. The information of the patients was as follows: (I) 70% patients were male, and the average age was 57 [27–85]; (II) according to TNM-staging system of gastric cancers, we used stage 0–I as early gastric cancer (n=60), and stage II–IV as advance gastric cancer; (III) normal gastric tissue samples were randomly selected from the patients (Gastritis, gastric ulcer, etc.) as normal controls. Tumor tissues, normal gastric tissue were extracted by the method of collagenase IV (0.1%) digestion of the tissue samples. The cell purity was determined by immunohistochemical staining with antibodies against the epithelial cytokeratins CK8 and CK18 (19).
Acquisition of specific aptamers
For pretreatment of the aptamer library (synthesized by Shanghai Biological Engineering Technology Co. Ltd., China), 1 nmoL of the aptamer library (5′-TAGATTGCACTTACTATC-54nt-ATTGAATAAGCTGGTATA-3′) was dissolved in 400 µL of binding buffer and denatured in boiling water for 5 min. The aptamers were then cooled on ice for 10 min, and 18 µL of bovine serum albumin (3%) was added to eliminate nonspecific binding.
Tissue cells were pretreated by mixing 1×106 early gastric cancer cells with the above-mentioned pretreated aptamer solution and set in an incubation (37 °C) for 30 min. After centrifugation at 100 ×g for 5 min, the mixture was washed twice with binding buffer to remove any unbound or weakly bound ssDNA. Two hundred microliters of ddH2O was added, and the bound aptamers were heated in a boiling incubation for 5 min. After centrifugation, the supernatant was collected.
The ssDNA was then dissolved in 20 µL of 1× TE binding buffer and the concentration was determined with a trace nucleic acid protein analyzer. The ssDNA was stored at −20 °C until use.
The obtained ssDNA was then used as a template for PCR amplification on the downstream biotin-labeled primer (see next section) to prepare the ssDNA library. Fifteen thermal cycles were conducted at 95 °C for 30 sec, 46.8 °C for 60 sec, and 72 °C for 30 sec. After purifying the PCR products, biotin-labeled ssDNA was obtained using biotin-streptavidin-coated magnetic beads (Promega, Madison, WI, USA) in order to prepare the secondary library. The amount of DNA was measured for the next round of selection. After each round of selection, filtering tests were applied to increase the specificity of cell-SELEX selection using normal gastric mucosa epithelial cells in order to remove any non-specific aptamers; i.e., those that could also bind to the normal gastric mucosa. The procedures described above were repeated for 12 rounds of selection. After each round of selection, we measured the binding rate of early gastric cancer cells used fluorescein - labeled ssDNA library. Two hundred pmol of the FAM-ssDNA library were bound with cells from the early gastric cancer, using ssDNA without cell as a blank control group, and then we measured the fluorescence intensity which represents the binding ability of the aptamer. (Lumina, Thermo Scientific, Waltham, MA, USA). Each sample was measured three times.
The ssDNA obtained after 12 rounds of selection was amplified using PCR with unlabeled upstream and downstream P1 and P4 primers, respectively (P1: 5′-TAGATTGCACTTACTATC-3′, P4: 5′-ATTGAATAAGCTGGTATA-3′; synthesized by Shanghai Biological Engineering Technology Co. Ltd.). After gel extraction, the ssDNA was inserted into a pGEM-T vector (Promega, Madison, WI, USA) and transformed into Escherichia coli DH5α competent cells (preserved in our laboratory). After gene cloning and blue-white selection, 26 positive clone colonies were chosen randomly and sent to Shanghai Biological Engineering Technology Co., Ltd. for sequencing.
Analysis of aptamer specificity and affinity
Plasmids were extracted from the 26 tubes of competent cells and amplified via PCR, using the upstream fluorescein (FAM)-labeled primer P2 (5′-FAM–TAGATTGCACTTACTATC-3′) and the downstream biotin-labeled primer P3 (5′-biotin–ATTGAATAAGCTGGTATA-3′), both synthesized at Shanghai Biological Engineering Technology Co., Ltd. After phenol: chloroform extraction, the aptamers were bound with streptavidin-coated magnetic beads in order to prepare fluorescein-labeled ssDNA.
To analyze specificity, 200 pmol of the aptamers were bound with cells from the early gastric cancer, advance gastric cancer and normal gastric mucosa samples, using aptamers without cell as a blank control group, and the fluorescence intensity was measured (Lumina, Thermo Scientific, Waltham, MA, USA). Each sample was measured three times.
To measure affinity, the fluorescein-labeled aptamers were diluted and bound with early gastric cancer cells. After washing and re-suspension, the fluorescent signal was measured (Lumina, Thermo Scientific). The average dissociation constants (Kd) of the 26 aptamers were analyzed according to the nonlinear fitting software (Origin 8.0): Y = Bmax × [X/(Kd + X)], where Bmax is the maximal binging capacity of the aptamer with target protein and X is a range of concentrations of aptamer, and Y is the fluorescence intensity of the test (11).
Extraction, purification, and identification of biomarkers of early gastric cancer
Tumor cells were obtained from 128 of the tissue specimens from patients with early gastric cancer described above by collagenase IV (0.1%) digestion. After purification, the cells were mixed in equal volume to obtain a mixed solution, and stored for later analysis.
Twenty µL of the bio-labeled aptamers (10 µmol/L) was placed in each centrifuge tube, and then 400 µL of binding buffer was added, followed by denaturing in boiling water for 5 min and cooling on ice for 10 min. The cell binding with the aptamer, 37 °C water bath for 20 min, 100 ×g centrifugal 5 min before washing. After the complex combined with magnetic beads (for 1 h at 4 °C), then cells were lysed. The supernatant was discarded on the magnetic rack before washing, binding with the elution buffer (for 20 min at 4 °C), the supernatant protein was stored at −20 °C, which were used for future research. SDS-PAGE and MALDI-TOF mass spectrometry analysis were used to get some information of the target proteins as described previously. After the above steps, we compared the protein sequence obtained by MALDI-TOF mass spectrometry against published sequences in the NCBI database using a BLAST search. We obtained protein hits showing a 100% match to the target protein and determined the information of the protein.
Evaluation of the new detection method for a candidate biomarker (EGA7)
To validate the method, the target protein obtained with the aptamer showing the highest specificity and affinity (Aptamer 7, Ap7) was designated EGA7, and was formulated into 13 different concentrations (600–3,000 ng/µL). Five microliters of the fluorescein-labeled aptamers (10 µmol/L) was placed in each centrifuge tube, and then 400 µL of binding buffer was added, followed by denaturing in boiling water for 5 min and cooling on ice for 10 min. Extracted EGA7 was mixed with the above solution and placed in an incubation (37 °C) for 30 min. After centrifugation at 1,600 ×g for 20 min, the supernatant was discarded, 400 µL of binding buffer was added, and the fluorescence intensity of each tube was measured; each sample was measured in triplicate.
The intra-assay reproducibility test was performed by measuring the fluorescence intensity of three samples on the same day 10 times to calculate the random error of the method. The inter-assay reproducibility test was performed by detecting the fluorescence intensity of three samples collected once a day for 10 days.
To determine the sensitivity of the detection method, the new biomarker (EGA7) was diluted to various concentrations (200–1,000 ng/µL). The EGA7 solutions were combined with each aptamer and the fluorescence intensity was measured. Each solution was tested three times. The data are expressed as the mean and standard deviation.
To determine the lowest effective concentration of aptamer required in the method, EGA solution at 3,000 ng/µL was used as the test sample. A series of concentrations of Ap7 solutions (1–14 µmol/L) was formulated, and the fluorescence intensity was recorded via fluorescence photometric analysis.
To evaluate and compare the binding ability of the aptamer with early gastric cancer cells, advanced gastric cancer cells, and normal gastric cells, 10 µmol/L FAM-Ap7 solution was added to each type of sample, and the fluorescence intensities were measured. The data were statistically analyzed with the independent-Sample T test using SPSS software.
Verification the scientific and reliability of the new detection method for a candidate biomarker (EGA7) via ELISA
In ELISA, the EGA7, which was extracted from single samples, was diluted twice by sample dilution buffer [0.05 mM Phosphate Buffer solution, PBS (pH 9.6)]. Wells of the ELISA plate (Corning) were coated overnight at 4°C with the EGA7. The follow day, wells were washed three times at room temperature with the wash solution [PBST (pH 7.4), PBS (pH 9.6) containing 0.1% (vol/vol) Tween 20]. Then it was blocked by 0.1% BSA buffer (for 4 h at 37 °C). After washing, EGA7 was detected (for 1 h at 37 °C) by ab16943 (Abcam, Cambridge, UK). After washing, bound antibody was detected by a goat-anti-mouse peroxidase (company) diluted (1:1000) before a 1 h incubation at 37 °C. The reaction was developed and test. The data were assayed with the independent-Sample T test using SPSS software.
Results
Identification of early gastric cancer cells
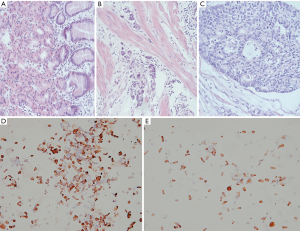
After the clinical diagnosis of early gastric cancer, specimens were separated with 0.1% collagenase type IV. Immunohistochemistry showed that the epithelial cells accounted for more than 95% of the specimens, which met the requirements for cell-SELEX screening (Figure 1).
Early gastric cancer cell aptamer screening and analysis of affinity specificity
After 12 rounds of cell-SELEX screening, the higher-affinity ssDNA aptamers in the library were retained. According to the optimal conditions for PCR amplification, all of the electrophoresis bands were close to 90 bp in length, suggesting that the obtained aptamers were ideally suited (Figure 2A).
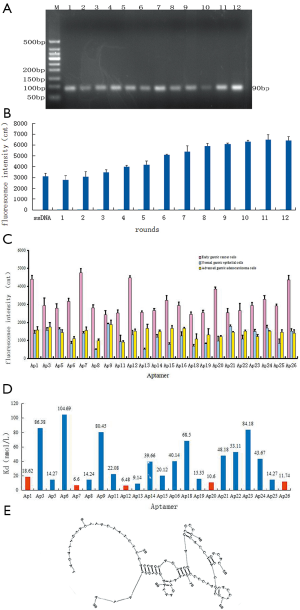
Fluorescence intensity was measured to reflect the capacity of the aptamers screened in each round to bind with the early gastric cancer cells (Figure 2B). The fluorescence intensity of the aptamers screened in the 12th round that bound to early gastric cancer cells was approximately twice that of the aptamers screened in the 1st round. High-affinity aptamers were enriched after the 10th round of screening, given that the fluorescence intensity value was essentially stable. Sequencing analysis showed that 4 of the 26 aptamers were inappropriate; specifically, Ap4, Ap10, and Ap17 showed notable double peaks, and Ap2 was 10 bp shorter than expected. Therefore, 22 apposite aptamers were selected for further analysis.
The specificity analysis showed the fluorescein-labeled aptamers that bound to early gastric cancer cells, normal gastric epithelial cells, and advanced gastric cancer cells, respectively (Figure 2C). The fluorescence intensity of the early-stage gastric cancer samples was significantly higher than that of the advanced gastric cancer and normal gastric mucosa samples (P<0.01). In addition, the fluorescence intensity of the advanced gastric cancer samples was also significantly higher than that of the normal gastric mucosa (P<0.01).
The affinity of aptamers was determined based on the histogram of the fluorescence intensity values reflecting the variation in binding ability of each aptamer to the early gastric cancer cells (Figure 2D). Specifically, a smaller dissociation constant (Kd) reflects greater affinity of the aptamers. Thus, the specificity and affinity analysis revealed that Ap1, Ap7, Ap12, Ap20, and Ap26 showed high specificity and affinity. Among them, Ap7 showed the highest specificity and affinity. The secondary structure of Ap7 is shown in Figure 2E.
Extraction, purification, and identification of EGA7
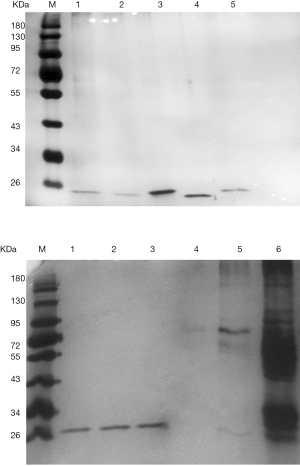
After extracting the target proteins, SDS-PAGE analysis revealed a single and clear electrophoretic band (Figure 3), and MALDI-TOF mass spectrometry provided detailed information of the target protein EGA7. Based on the mass spectrometry analysis and an NCBI database search, EGA7 was identified as peroxiredoxin-4 (Prx4). The nominal mass (Mr) was determined to be 30749 Da, pI was 5.86, with 271 amino acids; the best matched protein hit was to the reference sequence PRDX4_HUMAN.
Evaluation of the new biomarker (EGA7) detection method
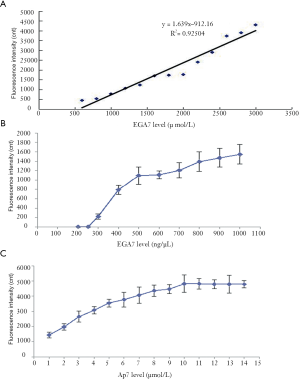
A linear regression was conducted according to the results of the binding ability between EGA7 and Ap7 (i.e., fluorescence intensity data). The linear regression obtained was Y = 1.6390X – 912.1 (R2 =0.9504), and the linear range of EGA7 was 556.53–3,000 ng/µL (Figure 4A).
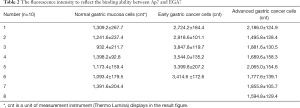
Because this is a new method, there is no threshold for comparison to determine accuracy; therefore, we compared the results based on imprecision thresholds of the Architect and ARK assays, which are <6.3% and <9.5%, respectively (20). The coefficient of variation for the intra-assay reproducibility was less than 6.5%, whereas the inter-assay coefficient of variation was larger than 9.5% (Table 1), suggesting that the new evaluation method is reproducible and the results are reliable.

Full table
The detection sensitivity analysis showed that the EGA7 concentration and fluorescence intensity values were positively correlated (Figure 4B). When the EGA7 concentration reached 300 ng/µL, the fluorescence intensity value was at the minimum. Therefore, the sensitivity of the detection method was deemed to be 300 ng/µL and the detection method showed a low threshold value.
A concentration of 3,000 ng/µL EGA7 which was the upper limit concentration of the linear regression equation was used as the test sample. A series of concentrations of Ap7 solutions was formulated, and the fluorescence intensity values (i.e., binding ability between EGA7 and the different concentrations of Ap7) were compared (Figure 4C). The minimum concentration of the curve platform (10 µmol/L) was acceptable for further testing.
The binding ability between Ap7 and EGA7 on the surface of different cells was also statistically compared according to variations in fluorescence intensity (Table 2), and significant differences among early gastric cancer cells, advanced gastric cancer cells, and normal gastric mucosa cells were noted (P<0.01).
Full table
Methods identification results
The result of ELISA (Figure 5) show that Prx4 in the early-stage gastric cancer was significantly higher than that in the advanced gastric cancer and normal gastric mucosa. In addition, Prx4 in the advanced gastric cancer was higher than the normal gastric mucosa. So the experimental results show that the new method is fully reliable.

Discussions
In a related study, aptamers were selected by cell-SELEX using advanced gastric cancer cells and tumor cell lines as target cells; however, these aptamers could not bind with early gastric cancer cells v (21). In order to avoid this problem, in the present study, all of the aptamers were selected using early gastric cancer cells as the target cells. Furthermore, to overcome the difficulty in obtaining research materials and to avoid selecting aptamers using only advanced gastric cancer cells and tumor cell lines as target cells, which are not suitable for capturing biomarkers from early gastric cancer cells, our study was carried out in collaboration with the People’s Hospital of Wuwei City and the Department of General Surgery of the Second Hospital of Lanzhou University. This ensured that all of the tumor specimens were obtained and extracted from patients with early-stage gastric cancer, and were identified using clinical and pathological analyses. After purification, the cells were mixed in equal volumes to obtain a mixed cell. This process was used to dilute specific substances reflecting individual characteristics, in order to eliminate the influence of individual differences as much as possible. The use of mixed cells from early tumor cells from different individuals allowed for specific aptamers to be obtained by selection, as the diversity among these individuals was discarded. A high-specificity aptamer would only bind with common substances which existing in the early gastric cancer cells, and should therefore reflect the specific expression characteristics of early tumor cells (22).
The MALDI-TOF mass spectrometry and NCBI database search showed that the target protein identified with the highest specificity and affinity, was Prx4. Therefore, the function of Prx4 is likely related to redox regulation of the cell (23,24). Prx4 may be subsequently regenerated by reduction of the disulfide bond via thioredoxin (25). There is no report of the use of Prx4 as a biomarker for the diagnosis of early-stage gastric cancer before. Therefore, Prx4 shows potential to be a novel biomarker for this purpose. However, further experiments are needed to examine whether the expression of the mRNA is consistent with the results of EGA7 expression determined via agarose gel electrophoresis.
The identification of Prx4 as a promising biomarker is specifically expressed in early gastric cancer cells shows good potential for reducing the mortality rate. One current limitation of this method is that Prx4 may only be a useful cancer biomarker at the tissue level, and gastric cancer tissue samples are difficult to obtain, especially for early-stage screening. Therefore, it would be useful to further determine whether Prx4 also exists in serum, and our next plan is analyzing the serum expression levels of Prx4 in gastric cancer patients. Further understanding of the mechanism of the regulation of Prx4 expression at the cell level may provide a deeper understanding of the development and pathogenesis of gastric cancer.
Acknowledgments
We are grateful to the Department of General Surgery, People’s Hospital of Wuwei City, Gansu Province, China, for providing the resected gastric cancer specimens. Funding: This work was supported by the National Natural Science Foundation of China (No.21272107); and the province science and technology support program (1304FKCA088).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.03.72). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All participants provided informed written consent for inclusion in this study. The study was approved by the School of Basic Medical Ethics Committee, Lanzhou University. Patients who underwent radical operation for the treatment of early gastric cancer at the People’s Hospital of Wuwei City and the Department of General Surgery of the Second Hospital of Lanzhou University were selected for inclusion.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Maehara Y, Kakeji Y, Oda S, et al. Tumor growth patterns and biological characteristics of early gastric carcinoma. Oncology 2001;61:102-12. [Crossref] [PubMed]
- Kim DH, Oh SJ, Oh CA, et al. The relationships between perioperative CEA, CA 19-9, and CA 72-4 and recurrence in gastric cancer patients after curative radical gastrectomy. J Surg Oncol 2011;104:585-91. [Crossref] [PubMed]
- Shimada H, Noie T, Ohashi M, et al. Clinical significance of serum tumor markers for gastric cancer: a systematic review of literature by the Task Force of the Japanese Gastric Cancer Association. Gastric Cancer 2014;17:26-33. [Crossref] [PubMed]
- Leung WK, Wu MS, Kakugawa Y, et al. Screening for gastric cancer in Asia: current evidence and practice. Lancet Oncol 2008;9:279-87. [Crossref] [PubMed]
- Zheng X, Hu B, Gao SX, et al. A saxitoxin-binding aptamer with higher affinity and inhibitory activity optimized by rational site-directed mutagenesis and truncation. Toxicon 2015;101:41-7. [Crossref] [PubMed]
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990;249:505-10. [Crossref] [PubMed]
- Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature 1990;346:818-22. [Crossref] [PubMed]
- Jarosch F, Buchner K, Klussmann S. In vitro selection using a dual RNA library that allows primerless selection. Nucleic Acids Res 2006;34:e86 [Crossref] [PubMed]
- Shangguan J, Li Y, He D, et al. A combination of positive dielectrophoresis driven on-line enrichment and aptamer-fluorescent silica nanoparticle label for rapid and sensitive detection of Staphylococcus aureus. Analyst 2015;140:4489-97. [Crossref] [PubMed]
- Gao L, Li Q, Li R, et al. Highly sensitive detection for proteins using graphene oxide-aptamer based sensors. Nanoscale 2015;7:10903-7. [Crossref] [PubMed]
- Lee CH, Han SR, Lee SW. Therapeutic Applications of Aptamer-Based Riboswitches. Nucleic Acid Ther 2016;26:44-51. [Crossref] [PubMed]
- Lorger M, Engstler M, Homann M, et al. Targeting the variable surface of African trypanosomes with variant surface glycoprotein-specific, serum-stable RNA aptamers. Eukaryot Cell 2003;2:84-94. [Crossref] [PubMed]
- Wang W, Chen C, Li X, et al. A bioresponsive controlled-release bioassay based on aptamer-gated Au nanocages and its application in living cells. Chem Commun (Camb) 2015;51:9109-12. [Crossref] [PubMed]
- Fang X, Tan W. Aptamers generated from cell-SELEX for molecular medicine: a chemical biology approach. Acc Chem Res 2010;43:48-57. [Crossref] [PubMed]
- Sefah K, Tang ZW, Shangguan DH, et al. Molecular recognition of acute myeloid leukemia using aptamers. Leukemia 2009;23:235-44. [Crossref] [PubMed]
- Sefah K, Meng L, Lopez-Colon D, et al. DNA aptamers as molecular probes for colorectal cancer study. PLoS One 2010;5:e14269 [Crossref] [PubMed]
- Kristian SA, Hwang JH, Hall B, et al. Retargeting pre-existing human antibodies to a bacterial pathogen with an alpha-Gal conjugated aptamer. J Mol Med (Berl) 2015;93:619-31. [Crossref] [PubMed]
- Linder S. Cytokeratin markers come of age. Tumour Biol 2007;28:189-95. [Crossref] [PubMed]
- Na W, Liu X, Wang L, et al. Label-free aptamer biosensor for selective detection of thrombin. Anal Chim Acta 2015;899:85-90. [Crossref] [PubMed]
- Zeng Y, Zheng A, Wu J, et al. Horseradish peroxidase and aptamer dual-functionalized nanoprobe for the amplification detection of alpha-methylacyl-CoA racemase. Anal Chim Acta 2015;899:100-5. [Crossref] [PubMed]
- Dàvila E, Parés D, Cuvelier G, et al. Heat-induced gelation of porcine blood plasma proteins as affected by pH. Meat Sci 2007;76:216-25. [Crossref] [PubMed]
- Shangguan D, Cao Z, Meng L, et al. Cell-specific aptamer probes for membrane protein elucidation in cancer cells. J Proteome Res 2008;7:2133-9. [Crossref] [PubMed]
- Eblimit A, Nguyen TM, Chen Y, et al. Spata7 is a retinal ciliopathy gene critical for correct RPGRIP1 localization and protein trafficking in the retina. Hum Mol Genet 2015;24:1584-601. [Crossref] [PubMed]
- Schrand B, Berezhnoy A, Brenneman R, et al. Reducing toxicity of 4-1BB costimulation: targeting 4-1BB ligands to the tumor stroma with bi-specific aptamer conjugates. Oncoimmunology 2015;4:e970918 [Crossref] [PubMed]


