
The mutational signatures and molecular alterations of bladder cancer
Introduction
Bladder cancer is the ninth most common cancer in the world, 430,000 new cases and 165,000 deaths each year around the world (1). The incidence is four times higher in men than in women; therefore, it is the seventh most common cancer in men and the seventeenth in women (2). The distribution of this type of cancer around the world differs (2). The highest incidences of bladder cancer are in Northern America and Europe (highest in Belgium, 17.5 cases per 100,000; age-standardized rate), while the lowest are in Asia, Latin America and Caribbean (i.e., 5.2 cases per 100,000 in South Korea). In Western countries, >90% of bladder cancers are transitional cell carcinoma (TCC) (urothelial carcinoma). However, in some developing countries, ~75% of cases are squamous cell carcinoma (3). These differences are presumably due to different carcinogenic exposures in different countries. Tobacco smoking and occupational exposures to aromatic amines and polycyclic hydrocarbons are the major risk factors in Western countries (4,5). However, in other countries, other environmental factors, i.e. schistosomiasis infections (Africa and the Middle East, i.e., Egypt), drinking water containing arsenic (Southeast Taiwan) and, presumably, aristolochic acid are responsible for cases of bladder cancer (6-8).
All cancers are transformed from normal cells as a result of somatic alterations in the genomes (9). Therefore, the landscape of somatic mutations in cancers provides deep insight into their pathophysiology, which is essential for designing efficient therapeutic modalities. During the last decade, more than thirty thousand human cancer tissues have been sequenced as an exome and/or whole genome together with their normal counterparts using massively parallel sequencing (or next-generation sequencing) technologies (10). Two large consortia, i.e., the International Cancer Genome Consortium (ICGC) and The Cancer Genome Atlas (TCGA), have led cancer genome studies worldwide and have advanced our understanding of cancer genomes (11,12). The development of many bioinformatics algorithms has enabled the identification of many types of somatic alterations in the genome, i.e., point mutations, copy number variations and chromosomal rearrangements, from the vast dataset now available (13). Several papers have reported the landscape of somatic mutations from large cohorts of bladder cancers, mostly from TCCs (14-19).
In this review, I summarize the features somatic mutations in the genomes of bladder cancer, mostly from TCCs, after which I provide an overview of genes and pathways frequently mutated in the cancer types. Finally, subtypes of bladder cancers classified by somatic mutations are briefly introduced.
Mutational signatures and processes
Like all cells that constitute an individual, a cancer cell is a direct descendant of the fertilized egg. As a result, excluding very small exceptions (i.e., DNA from a virus), genomes in the cancer cell were inherited from the fertilized egg through a lineage of mitotic cell divisions. The accuracy of the DNA replication process is very high but not perfect. Therefore, somatic mutations are acquired during every cell division due to the slight infidelity of the intrinsic DNA replication machinery (9). In addition, incidences of exposure to exogenous and/or endogenous mutagens (e.g., ultraviolet light and tobacco smoking in skin cancers and lung cancers, respectively), enzymatic modification of DNA (e.g., APOBEC-related mutagenesis frequently observed in breast cancers), and defective DNA repair (e.g., defective mismatch repairs in colorectal cancer cases) are also responsible for somatic mutations (20). Somatic mutations are archeological traces of previous cell divisions left in cancer cells; for this reason, they can provide deep insight into the mutational processes that have operated in the somatic cell lineages from the fertilized egg (21).
From the genomes of bladder cancers, on average, we find approximately eight somatic base substitutions per megabase (Mb) (14,15,20,22). Given the sizes of whole genomes (2,950 Mb) and protein-coding regions (approximately 40 Mb), we expect approximately 24,000 genome-wide and 300 coding-sequence mutations. However, mutation rates vary extensively within bladder cancer patients (14,15,20,22). The frequency ranges from 1 to 60 per Mb (this broad range is discussed in more detail below). Compared to other common cancer types, the average rate of somatic base substitutions in bladder cancers is surprisingly high; only melanoma, lung squamous and lung adenocarcinoma show higher average mutation rates. Bladder cancer shows a higher rate than small cell carcinomas of the lung and many other solid cancers, such as cancers of the colorectal, stomach, liver, prostate, breast and pancreas. It is important to note that a substantial proportion of somatic mutations in the three types of cancer tissues with higher mutation rates than bladder cancers are attributable to exposure to well-known, strong carcinogens, i.e., ultraviolet light (melanoma) and tobacco smoke (lung squamous and adenocarcinoma) (20).
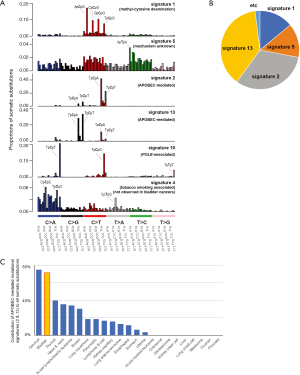
Each mutational process imprints a distinct signature of mutations onto the genomes on which it has been operative (20,23,24). For example, tobacco smoke generates mainly C:G > A:T substitutions while ultraviolet light leaves predominantly C:G > T:A substitutions at dipyrimidine contexts. Several groups have developed methods by which to extract mutational signatures from human cancers employing a 96-base substitution classification based on six base substitution classes (i.e., C > A, C > G, C > T, T > A, T > C, T > G; all substitution types are referred to by the pyrimidines of the mutated Watson-Crick base pairs) and the immediate 5' and 3' sequence contexts of each mutated base (20,23). As a result, more than 30 different mutational signatures have been identified (an up-to-date catalogue of signatures is available at COSMIC database, http://cancer.sanger.ac.uk/cosmic/signatures). Interestingly, in the hundreds of genomes of bladder cancers, only five mutational signatures, i.e., signatures 1, 2, 5, 10 and 13, have been observed (Figure 1A; Table 1; see COSMIC database for more signatures) (20).

Full table
Of these, signatures 1 and 5 explain approximately 30% of somatic mutations (~15% each) observed in cases of bladder cancer (Figure 1A,B) (14-16,20). Neither signature, however, is specific to bladder cancer, but both are frequently seen in most cancer samples in all cancer types (25). Signature 1 is thought to be due to the spontaneous deamination of 5-methyl cytosine to thymine, which results in C > T transitions primarily at the CpG dinucleotides. Signature 5 primarily features C > T and T > C transitions, but its underlying biology is currently unknown. In bladder cancers, signature 5 is associated with somatic ERCC2 mutations (26). Generally, the numbers of mutations attributable to both signatures exhibit strong positive correlations with age upon the diagnosis of cancer (25). Furthermore, signatures 1 and 5 appear to be the dominant mutational signatures contributing to human early embryonic mutations and de novo germline mutations (32,33). Taken together, the underlying processes of these two signatures are endogenous and are universally operative in most normal somatic cells, including cells in the bladder, at relatively constant rates throughout life (25).
Signature 10 has been found in a very small fraction of bladder cancer cases (Figure 1A). This signature is associated with altered activity of error-prone DNA polymerase POLE and causes primarily C > A and C > T substitutions at the TpCpT context and T > G mutations at the TpTpT context (20). This signature has been frequently found in ultra-hypermutators (i.e., mutation rate >30 per Mb) of colorectal and uterine cancers (29,30). However, the ultra-hypermutators in bladder cancers do not always show this signature (20); therefore, its contribution in cases of bladder cancer is considered to be relatively minimal compared to that in colorectal and uterine cancers (Figure 1B).
The most remarkable mutational signatures contributing to the genomes of bladder cancers are signatures 2 and 13, explaining ~70% of somatic mutations (Figure 1A,B) (20). These signatures are found more than 70% of bladder cancer samples. Signatures 2 and 13, characterized by C > T transitions and C > G transversions at the TpCpA and TpCpT motifs, respectively, are attributed to APOBEC-mediated mutagenesis coupled to activity of the base excision repair and DNA replication machineries (24). Among 11 members of the AID/APOBEC enzymes (34), APOBEC3A and/or APOBEC3B likely underlie the signature 2 and 13 mutations (20,27,28). Generally, APOBEC activation constitutes part of the innate immune response to viruses and retrotransposons. However, the correlation between the virus/retrotransposon activation and APOBEC signatures is not remarkable in cancer samples; therefore, the reason for the APOBEC-mediated mutagenesis in somatic cells is not clear. These mutational signatures were initially identified in breast cancer genomes (24) but are found in >20 cancer types, including non-small-cell lung, uterine, stomach, pancreas and cervical cancers. Except for cervical cancer, the dominant contribution of APOBEC in somatic mutations (i.e., attributable to ~70% somatic mutations in bladder cancer) is not observed in other cancer types (Figure 1C; i.e., ~30% in breast cancer, ~10% in non-small-cell lung cancer) (20,24). In many cancer types, such as breast and non-small-cell lung cancers, APOBEC-mediated mutations are occasionally highly enriched in the localized genomic regions (termed kataegis) (24). Despite the hyperactivity of APOBEC, kataegis is not frequently observed in bladder cancers (20).
Tobacco smoking is a well-known risk factor for bladder cancer. The age-adjusted odds ratio for current smokers to develop bladder cancer is as high as 4.0 (35). Signature 4 is directly associated with tobacco smoking, characterized mainly by C > A substitutions (Figure 1A) (20). Signature 4 is very similar to the mutation signature induced in vitro by exposing cells to benzo[a]pyrene, a major carcinogen in tobacco smoke (31). Signature 4 mutations are only found in cancer types in which tobacco smoking increases risk and mainly in those from epithelia directly exposed to tobacco smoke. In line with this, despite the known risk conferred by tobacco smoking (i.e., odds ratio =4.0) in bladder cancers, signature 4 mutations are not observed in the genomes (14,20). The total numbers of base substitutions and SVs are not significantly different between smokers and non-smokers in cases of bladder cancer. Instead, more signature 5 mutations are found in smokers compared to non-smokers in bladder cancer cases (31). It appears that tobacco smoking indirectly influences bladder cancer by activating the molecular machinery underlying signature 5 rather than directly damaging the genomic DNA (31).
Tobacco smoking may affect bladder cancer by inducing hyper-methylation of promoter CpG islands. The CpG island methylation phenotype (CIMP) may be more frequently observed in bladder cancers of smokers (14,36). However, genome-wide CpG methylation showed limited differences between the cancers of smokers and non-smokers (31).
Structural variations (SV)
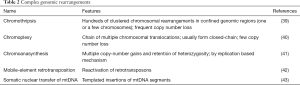
Somatic SV is defined as the entire repertoire of genome rearrangements accumulated through a lineage of mitotic cell divisions. Theoretically, SV can be acquired before or after cellular transformation from normal to cancer cells. Indeed, genome instability is a well-known hallmark of cancer cells (37) and a variety of SVs, including copy number alterations (CNAs) (i.e., large deletions and amplifications), intrachromosomal inversions and interchromosomal translocations, are frequently observed in cancer genomes (38). Classically, SVs have been revealed using cytogenetics technologies. Since 2008, our understanding of cancer SVs has been greatly enhanced due to the application of microarray and whole-genome sequencing, which enable the detection of SVs systematically at the nucleotide resolution. As a result, several new complex SV patterns and mechanisms have been identified, such as chromothripsis (39), chromoanasynthesis (40), chromoplexy (41), mobile-element retrotransposition (42) and nuclear-transfer of mitochondrial DNA (43), to name a few (Table 2). However, compared to the substitution mutations discussed above, the SV detection accuracy (sensitivity and specificity) is more affected by technical issues (i.e., the read-depth of sequencing and the bioinformatics algorithms applied). Therefore, unbiased comparisons of SVs across many cancer types have yet to be published. Ongoing large-scale cancer genome studies such as the ICGC Pan-Cancer Analysis of Whole Genomes (PCAWG) will provide a more sophisticated understanding (https://dcc.icgc.org/pcawg) (44).
Full table
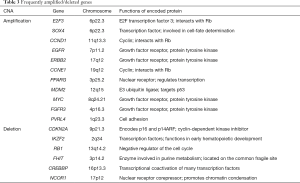
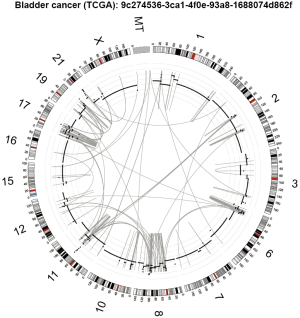
Generally, SVs are frequent in bladder cancers, and several genomic regions are recurrently amplified and deleted in many samples (Table 3) (14). For example, amplifications of 6p22.3 (E2F3 and SOX4), 11q13.3 (CCND1), 7p11.2 (EGFR), 17q12 (ERBB2), 19q12 (CCNE1), 3p25.2 (PPARG), 12q15 (MDM2), 8q24.21 (MYC), 4p16.3 (FGFR3) and 1q23.3 (PVRL4) and deletions of 9p21.3 (CDKN2A), 2q34 (IKZF2), 13q14.2 (RB1), 3p14.2 (FHIT), 16p13.3 (CREBBP) and 17p12 (NCOR1) are commonly identified. These CNAs are correlated with increased/reduced expressions of the genes involved, suggesting that alterations of these genes contribute to the development and/or maintenance of bladder cancers. Inversion- and translocation-type SVs are also quite common in bladder cancers. Overall, bladder cancers harbor ~300 SVs per sample (ICGC-PCAWG dataset; data not shown), ranging from 30 to 1,000. The SV rate in bladder cancer is higher than in many cancer types (i.e., colorectal, liver, pancreas and cervical cancers) and comparable to those associated with non-small-cell lung and uterine cancers. A proportion of these SVs observed in a sample are clustered and inter-connected, suggesting complex SV mechanisms (i.e., chromothripsis, chromoanasynthesis and chromoplexy) may be frequently operative in the somatic lineages of bladder cancer cells (Figure 2). Interestingly, the somatic nuclear transfer of mitochondrial DNA is also observed in bladder cancer (Figure 2), which has been frequently detected in non-small-cell lung cancers, melanoma, breast and uterine cancers but not in colorectal and stomach cancers. All of these features confirm that SVs are widespread in bladder cancer genomes.
Full table
As results, in a subset of bladder cancers (<10%), some in-frame fusion receptor tyrosine kinases (RTKs) are observed (14,15). FGFR3-TACC3 fusion genes are recurrently generated (~3% in bladder cancers) by intrachromosomal translocation (chromosome 4) (14,15,45). The kinase domain in the chimeric FGFR3 protein can be constitutively activated by auto-dimerization. This fusion gene is also reported in glioblastoma (46) thus is highly likely to have pathogenic significance and is a promising therapeutic target as many fusion genes (i.e., EML4-ALK and KIF5B-RET) in lung adenocarcinoma (47).
In the TCGA study, several viruses, such as cytomegalovirus (CMV), BK polyoma virus and human papilloma virus 16 (HPV16), have been identified in a small subset of bladder cancer samples (<10%) (14). Although CMVs were present as episomes in the samples, viral genes from the BK polyoma virus and HPV16 were integrated into nuclear genes (such as GRB14 and BCL2L1) and were expressed in the cancer cells. These findings suggest that viral infections may play a role in the development of a small proportion of bladder cancers.
Driver mutations and altered pathways
In cases of bladder cancer, a subset of protein-coding genes is significantly more frequently mutated than expected by chance (summarized in Table 4) (14,15,48). This suggests that these mutations are important for the carcinogenesis of bladder cancers.
Full table
Approximately 50% of bladder cancer samples harbor TP53 somatic mutations, which inactivate the TP53 functions (Figure 3A). MDM2 amplification and/or overexpression are observed in approximately 25% of samples without TP53 somatic mutations. Overall, the TP53 function is inactivated in 75% of bladder cancer cases. CDKN2A deletion and RB1 inactivating mutations are observed in approximately 50% and 13% of the samples, respectively. The two mutation types are mutually exclusive. In addition, focal amplifications of CCND1 (~10%), CCNE1 (~10%) and E2F3 (~20%) occur very frequently. RB1 and E2F3 alterations were reported to be more prevalent in the tumors exhibiting neuroendocrine differentiation (19). Overall, >90% of bladder cancer samples show dysregulation of the p53/RB cell cycle pathway (14).
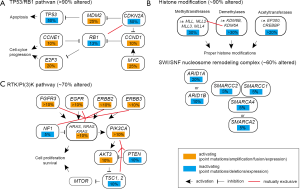
Chromatin remodeling pathways, including histone modification (~90%) and SWI/SNF nucleosome remodeling complex (~60%), are also frequently altered by somatic mutations (Figure 3B) (14,15,48). For histone modification, histone methyltransferases [including MLL, MLL2 (also known as KMT2D), MLL3, MLL4 and SETD2; overall >30%], histone demethylases (including KDM6A and KDM6B; overall >30%), and histone acetyltransferases (including CREBBP, EP300; overall >20%) are frequently inactivated. Mutations in MLL2 and KDM6A were found to be mutually exclusive, suggesting that the inactivation of those two genes has redundant downstream effects on carcinogenesis. With respect to the SWI/SNF nucleosome remodeling complex, ARID1A, ARID1B, SMARCA2, SMARCA4, SMARCC1 and SMARCC2 genes are frequently inactivated (14).
Alterations in the RTK and phosphatidylinositol-3-OH kinase [PI(3)K] pathways are observed in ~70% of bladder cancer samples (Figure 3C) (14,15,48). The RTK pathway is activated in ~40% of cancer samples preferentially with FGFR3 (activating point mutation, amplification, and FGFR3-TACC3 fusions; overall >10%), EGFR (mostly amplification; >10%), ERBB2 (activating mutation and amplification; ~10%), ERBB3 (activating point mutation; ~10%), NRAS/HRAS/KRAS (activating mutation; overall ~10%) and inactivation mutations of NF1 (~5%). These mutations are mutually exclusive. Likewise, the PI(3)K pathway is activated in ~40% of samples preferentially by PIK3CA (activating mutation; >10%), TSC1 and TSC2 (inactivating mutation and deletion; overall ~10%), PTEN (inactivating mutation; ~10%) and the overexpression of AKT3 (~10%).
Through large-scale genome sequencing studies, several genes in other pathways were recently identified as bladder cancer genes. Most interestingly, STAG2 (encoding stromal antigen 2) was observed to be inactivated in >10% of bladder cancers by truncating mutations, large deletions and promoter hyper-methylations (15). This gene encodes a protein associated with the sister chromatid cohesion and segregation process of the cell cycle. Segregation errors during mitosis usually result in micronuclei, where lagging chromosomes or chromatid fragments are clustered and DNA double-strand break repair processes, frequently error-prone, take place (49). Complex rearrangement mechanisms such as chromothripsis can be generated in micronuclei (50). Therefore, STAG2 gene inactivation may be associated with the high frequency of SVs in bladder cancers, consistent with previous reports of glioblastoma, melanoma and Ewing’s sarcoma (51). Clinically, bladder cancer patients with inactivated STAG2 showed significantly lower survival rates than wild-type individuals (15).
The ERCC2 gene, which encodes a DNA helicase that plays a central role in the nucleotide-excision repair pathway, is inactivated in ~10% bladder cancer samples by recurrent deleterious missense mutations (14,15). As noted above, ERCC2 mutant bladder cancer samples show higher numbers of somatic mutations attributable to mutation signature 5 (26), suggesting a role of this gene in shaping the mutational landscape of bladder cancers.
Retinoic X nuclear receptor alpha (RXRA) activating mutations are detected in ~10% of samples (14). The mutant samples activate a subset of genes involved in adipogenesis and lipid metabolism. In addition, ~10% of bladder cancer samples harbor deleterious missense mutations in NFE2L2 (14), a transcription factor that regulates the anti-oxidant program in response to oxidative stress. Similarly, TXNIP, which encodes thioredoxin-interacting protein, is inactivated in ~7% in a mutually exclusive manner to NFE2L2 (14). Mutations in the two genes suggest that dysregulation of the anti-oxidation pathway has a functional role in the pathogenesis of bladder cancers.
Finally, approximately 10% of bladder cancer samples have missense mutations in either RHOA or RHOB (14). These genes encode members of the Rho family of small GTPases, which are Ras-like proteins that act as intermediaries between cell surface receptors (52). The RHOA gene was recently identified as a novel cancer gene in diffuse-type gastric cancers (53).
Molecular stratification and therapeutic targets of bladder cancers
Using somatic point mutations and CNAs, studies involving unsupervised clustering found that there are three distinct subtypes in bladder cancers (Table 5) (14). The first group (group A) is characterized by “focal amplification,” where focal somatic CNAs in several genes (i.e., EGFR, PPARG, PVRL4, CCNE1, MYC and EGFR) are highly enriched. Cancer samples in this group also harbor high-frequency MLL2 mutations. The second group (group B) is characterized by “CDKN2A-deficient & FGFR3 mutants”. Losses of CDKN2A and FGFR3 activating mutations (i.e., point mutations, amplifications and instances of FGFR3-TACC3 fusion) are observed in the vast majority of the samples in this group. These samples frequently show a papillary histology. The last group (group C) is classified as “TP53/cell-cycle-mutants”, where TP53 mutations are observed in nearly all samples and further alterations in genes in the pathway (i.e., RB1, E2F3 and CCNE1) are enriched. These clusters suggest that the molecular mechanisms for bladder cancers are not homogeneous in many samples and that different therapeutic strategies may be necessary according to the molecular groups for more efficient treatments of bladder cancers.

Full table
Analysis of gene expression profiles suggests four clusters in bladder cancers (Table 5) (14). Cluster I is enriched in tumors with papillary histology, activating FGFR3 alterations (mutations, amplifications, FGFR3-TACC fusions and overexpression) and poor gene expression of miR-99a and miR-10 which down-regulate FGFR3 expression. Tumors clustered in this group may respond to FGFR inhibitors. Cluster I and II tumors show high mRNA and protein expression of GATA3, FOXA1, UPK3A, ERBB2 (HER2) and ESR2. HER2 protein levels in a subset of bladder cancers in clusters I and II are comparable to those found in HER2-positive breast cancers. These signatures indicate potential targets for human epidermal growth factor receptor inhibitors (HER2) and/or hormone therapies such as tamoxifen or raloxifen for bladder cancers in clusters I and II. Signature for bladder cancer cluster III is “basal/squamous-like”, similar to that of basal-like breast cancers. Cluster III is enriched in tumors with squamous histology, and higher expression of genes specific to the epithelial lineage, such as cytokeratins (i.e., KRT14, KRT5, KRT6A) and EGFR. Finally, cluster IV tumors do not show features mentioned above, including FGFR3 activation (cluster I), overexpression of GATA3, FOXA1, UPK3A, ERBB2, ESR2 (cluster I and II), and cytokeratins/EGFR (cluster III).
Bladder cancer genome studies identified several genomic and pathway alterations amenable to therapeutic targeting, including PI(3)K/AKT/mTOR (in ~40% of bladder cancers) and RTK (i.e., ERBB2, ERBB3 and FGFR3)/RAS (in ~40% of bladder cancers) pathways. These two pathways are therapeutic targets in other cancer types (i.e., breast and lung cancers). For PI(3)K/AKT/mTOR pathway alteration, PI(3)K inhibitors (for PIK3CA mutations), mTOR inhibitors (for TSC1 and TSC2 mutations) and AKT inhibitors (for AKT3 overexpression) can be considered. For RTK/RAS pathway alteration, FGFR inhibitors/antibodies (for FGFR3 alterations), EGFR antibodies/inhibitors (for EGFR amplification), ERBB kinase inhibitors (for ERBB3 mutation) and ERBB2 kinase inhibitors/antibodies (ERBB2 mutation and amplification) are potential therapeutic targets. In addition, the alterations in epigenetic pathways, altered in ~90% of bladder cancers, provide another avenue for bladder cancer treatment. Therapeutics that target chromatin modifications, i.e., recently developed bromodomain and extra-terminal (BET) inhibitors, maybe prove useful for treatment of bladder cancers with alterations in chromatin regulatory genes (54).
Future perspectives
Genome sequencing technologies provide opportunities for understanding the comprehensive molecular alterations in human cancers. To date, several hundred bladder cancers have been sequenced especially for TCCs. Clearly, more bladder cancer samples, including other histology types, will be sequenced in the futures. Clinically, it would be necessary to sort out genomic mutations which are clearly associated with drug response, metastasis, recurrence and prognosis. In line with this, cancer multi-region sequencing (an analysis of matched primary and metastatic tissues from the same patients) (55) and longitudinal sequencing (an analysis of matched early primary and late recurred tissues from the same patients) (56) will be the direction of future studies. Basically, it would be really interesting to reveal the reason and the timing of the active APOBEC-mediated mutagenesis through the life history of bladder cancer cells from the fertilized egg to the timing of cellular transformation. Remarkably, observation of the mutational processes in normal cells will address this possibility (57,58). These efforts will provide deep insights into the prevention of bladder cancers.
Acknowledgments
Funding: This work was supported by a grant of the Korea Health Technology R&D project through the Korea Health Industry Development Institute (KHIDI) funded by the Ministry of Health & Welfare, Republic of Korea (HI14C1277).
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Ja Hyeon Ku, Kunyoo Shin, Minyong Kang) for the series “Bladder Cancer” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.05.37). The series “Bladder Cancer” was commissioned by the editorial office without any funding or sponsorship. The author has no other conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108.
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86.
- Pakzad R, Mohammadian-Hafshejani A, Mohammadian M, et al. Incidence and Mortality of Bladder Cancer and their Relationship with Development in Asia. Asian Pac J Cancer Prev 2015;16:7365-74.
- Kogevinas M, 't Mannetje A, Cordier S, et al. Occupation and bladder cancer among men in Western Europe. Cancer Causes Control 2003;14:907-14.
- Letasiova S, Medve'ova A, Sovcikova A, et al. Bladder cancer, a review of the environmental risk factors. Environ Health 2012;11:S11.
- Poon SL, Huang MN, Choo Y, et al. Mutation signatures implicate aristolochic acid in bladder cancer development. Genome Med 2015;7:38.
- Chen CL, Chiou HY, Hsu LI, et al. Arsenic in drinking water and risk of urinary tract cancer: a follow-up study from northeastern Taiwan. Cancer Epidemiol Biomarkers Prev 2010;19:101-10.
- Kirkali Z, Chan T, Manoharan M, et al. Bladder cancer: epidemiology, staging and grading, and diagnosis. Urology 2005;66:4-34.
- Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature 2009;458:719-24.
- Garraway LA, Lander ES. Lessons from the cancer genome. Cell 2013;153:17-37.
- Nik-Zainal S, Davies H, Staaf J, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016;534:47-54.
- Farshidfar F, Zheng S, Gingras MC, et al. Integrative Genomic Analysis of Cholangiocarcinoma Identifies Distinct IDH-Mutant Molecular Profiles. Cell Rep 2017;18:2780-94.
- Tian R, Basu MK, Capriotti E. Computational methods and resources for the interpretation of genomic variants in cancer. BMC Genomics 2015;16:S7.
- . Cancer Genome Atlas Research N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014;507:315-22.
- Guo G, Sun X, Chen C, et al. Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Nat Genet 2013;45:1459-63.
- Cazier JB, Rao SR, McLean CM, et al. Whole-genome sequencing of bladder cancers reveals somatic CDKN1A mutations and clinicopathological associations with mutation burden. Nat Commun 2014;5:3756.
- Ross JS, Wang K, Al-Rohil RN, et al. Advanced urothelial carcinoma: next-generation sequencing reveals diverse genomic alterations and targets of therapy. Mod Pathol 2014;27:271-80.
- Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature 2013;502:333-9.
- Iyer G, Al-Ahmadie H, Schultz N, et al. Prevalence and co-occurrence of actionable genomic alterations in high-grade bladder cancer. J Clin Oncol 2013;31:3133-40.
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415-21.
- Alexandrov LB, Stratton MR. Mutational signatures: the patterns of somatic mutations hidden in cancer genomes. Curr Opin Genet Dev 2014;24:52-60.
- Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013;499:214-8.
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Deciphering signatures of mutational processes operative in human cancer. Cell Rep 2013;3:246-59.
- Nik-Zainal S, Alexandrov LB, Wedge DC, et al. Mutational processes molding the genomes of 21 breast cancers. Cell 2012;149:979-93.
- Alexandrov LB, Jones PH, Wedge DC, et al. Clock-like mutational processes in human somatic cells. Nat Genet 2015;47:1402-7.
- Kim J, Mouw KW, Polak P, et al. Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nat Genet 2016;48:600-6.
- Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet 2013;45:977-83.
- Roberts SA, Lawrence MS, Klimczak LJ, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet 2013;45:970-6.
- Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330-7.
- Cancer Genome Atlas Research N. Integrated genomic characterization of endometrial carcinoma. Nature 2013;497:67-73.
- Alexandrov LB, Ju YS, Haase K, et al. Mutational signatures associated with tobacco smoking in human cancer. Science 2016;354:618-22.
- Rahbari R, Wuster A, Lindsay SJ, et al. Timing, rates and spectra of human germline mutation. Nat Genet 2016;48:126-33.
- Ju YS, Martincorena I, Gerstung M, et al. Somatic mutations reveal asymmetric cellular dynamics in the early human embryo. Nature 2017;543:714-8.
- Salter JD, Bennett RP, Smith HC. The APOBEC Protein Family: United by Structure, Divergent in Function. Trends Biochem Sci 2016;41:578-94.
- Freedman ND, Silverman DT, Hollenbeck AR, et al. Association between smoking and risk of bladder cancer among men and women. JAMA 2011;306:737-45.
- Samowitz WS, Albertsen H, Sweeney C, et al. Association of smoking, CpG island methylator phenotype, and V600E BRAF mutations in colon cancer. J Natl Cancer Inst 2006;98:1731-8.
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74.
- Tubio JM. Somatic structural variation and cancer. Brief Funct Genomics 2015;14:339-51.
- Stephens PJ, Greenman CD, Fu B, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 2011;144:27-40.
- Holland AJ, Cleveland DW. Chromoanagenesis and cancer: mechanisms and consequences of localized, complex chromosomal rearrangements. Nat Med 2012;18:1630-8.
- Baca SC, Prandi D, Lawrence MS, et al. Punctuated evolution of prostate cancer genomes. Cell 2013;153:666-77.
- Tubio JM, Li Y, Ju YS, et al. Mobile DNA in cancer. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science 2014;345:1251343
- Ju YS, Tubio JM, Mifsud W, et al. Frequent somatic transfer of mitochondrial DNA into the nuclear genome of human cancer cells. Genome Res 2015;25:814-24.
- Stein LD, Knoppers BM, Campbell P, et al. Data analysis: Create a cloud commons. Nature 2015;523:149-51.
- Williams SV, Hurst CD, Knowles MA. Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet 2013;22:795-803.
- Singh D, Chan JM, Zoppoli P, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science 2012;337:1231-5.
- Seo JS, Ju YS, Lee WC, et al. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res 2012;22:2109-19.
- Knowles MA, Hurst CD. Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nat Rev Cancer 2015;15:25-41.
- Fenech M, Kirsch-Volders M, Natarajan AT, et al. Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis 2011;26:125-32.
- Zhang CZ, Spektor A, Cornils H, et al. Chromothripsis from DNA damage in micronuclei. Nature 2015;522:179-84.
- Solomon DA, Kim T, Diaz-Martinez LA, et al. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science 2011;333:1039-43.
- Wang HR, Zhang Y, Ozdamar B, et al. Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science 2003;302:1775-9.
- Kakiuchi M, Nishizawa T, Ueda H, et al. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat Genet 2014;46:583-7.
- Filippakopoulos P, Knapp S. Targeting bromodomains: epigenetic readers of lysine acetylation. Nat Rev Drug Discov 2014;13:337-56.
- Gundem G, Van Loo P, Kremeyer B, et al. The evolutionary history of lethal metastatic prostate cancer. Nature 2015;520:353-7.
- Bolli N, Avet-Loiseau H, Wedge DC, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun 2014;5:2997.
- Blokzijl F, de Ligt J, Jager M, et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature 2016;538:260-4.
- Martincorena I, Roshan A, Gerstung M, et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 2015;348:880-6.


