
Identification of new biomarkers for cisplatin-resistance in A549 human lung adenocarcinoma cells by an integrated bioinformatical analysis
Introduction
Lung cancer has the highest incidence and causes the highest mortality among all cancers in China, which was projected to strike about 733,300 Chinese and to cause almost 610,200 cancer-related mortality in 2015 (1). Although more and more drugs have been developed and applied to treat lung cancer, the therapeutic efficacy is still poor. Platinum chemotherapeutic agents, such as cisplatin, are conventional drugs for treating advanced lung cancer. Despite the favorable initial response, they finally become inefficient in most patients because of cisplatin resistance (2).
Recently, various new molecular biomarkers have been discovered to overcome the drug resistance. For instance, the protein disulfide isomerases PDIA4 and PDIA6 mediate the resistance to cisplatin-induced cell death in lung adenocarcinoma (3). The copper transporter CTR1 can regulate the uptake of cisplatin and has been developed as a therapeutic target (4). The multidrug resistance protein MRP2 can increase the efflux of cisplatin and is a biomarker for cisplatin resistance in advanced esophageal squamous cell carcinoma patients (5,6). Medical treatments should be designed according to the characteristics of each patient. It is necessary to discover more effective biomarkers and reveal the underlying molecular mechanisms of cisplatin resistance.
Microarray is a powerful tool for detecting gene expression pattern, including those mRNA and microRNAs (miRNAs). Due to the wide use of microarray, a large number of microarray data have been collected. Bioinformatical methods are crucial for discovering more valuable information contained in these datasets, particularly those of signaling pathways, complex biological processes and the interaction network of differentially expressed genes (DEGs).
In this study, two microarray datasets, one mRNA dataset (E-MEXP-3123) and one miRNA dataset (GSE43249), were obtained from GEO and ArrayExpress database. The DEGs and differentially expressed miRNAs (DEMs) between parental and cisplatin-resistant A549 lung cancer cells were identified. We performed a functional enrichment analysis and analyzed PPI, miRNA network and their target genes. Some candidate biomarkers were tested by the cisplatin-resistance experiment in A549 cell lines.
Methods
Microarray data
GEO (https://www.ncbi.nlm.nih.gov/geo/) and ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) are two public functional genomics data repositories. They contain large free array and sequence-based datasets. E-MEXP-3123 is an expression profiling of platinum drugs resistant genes in A549 cell lines, including three wild A549 cell samples and three cisplatin-resistant A549 cell samples. GSE43249 is a miRNA expression profiling of cisplatin-resistant cells derived from the A549 lung cell line. It also contained the data of six samples, including those of three A549 cell samples and three cisplatin-resistant A549 cell samples.
Identification of DEGs and DEMs
R (version 3.3.2) is a free software environment for statistical computing and graphics. Data processing was accomplished by using the R package limma, and the DEGs and DEMs were identified between parental and cisplatin-resistant lung cancer cells. Empirical Bayes statistics and Benjamini-Hochberg correction were applied to control the false-positive results. Expression change more than 2 folds (≥2 folds) and the adjusted P values (adj. P.val) less than 0.05 (<0.05) denoted a statistical difference. A heat map of DEGs was generated by using the R package pheatmap, and the volcano plot of DEMs was drawn by using the R package ggplot2.
Functional analysis and pathway enrichment analysis
DAVID (http://david.ncifcrf.gov/) is an online tool that can be utilized to perform functional analysis and pathway enrichment analysis for discovering the relationships among the selected gene sets (7). Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed by the DAVID online program. The R package ggplot2 was employed to present the related results.
PPI network construction and module analysis
All DEGs were uploaded to the Search Tool for the Retrieval of Interacting Genes (STRING, version 10.0, http://string.embl.de/) database which collected the data of 9.6 million proteins and 184 million interactions from 2,031 organisms (8). The combined score ≥0.7 was set as the cut-off criterion. Cytoscape (version 3.4.0) was then employed to generate a PPI network (9). The significant modules were selected by Molecular Complex Detection (MCODE) which is an app at the Cytoscape store (10). The advanced options were set as follows: degree cutoff =2, node score cutoff =0.2, k-core =2, max-depth =100.
Analysis of miRNA-target mRNA network
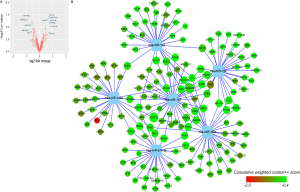
The top 6 DEMs were imported to TargetScan (Release 7.1, http://www.targetscan.org/) which can search the target genes based on the conserved sites matching the seed regions of miRNAs. Cumulative weighted context++ score ≤−0.4 were set as the threshold. The miRNA-target mRNA network was constructed by Cytoscape.
Cell culture and cell viability assay
Normal and cisplatin-resistant human A549 lung cells were obtained from Zhongshan University. The cells were cultured in minimal Roswell Parker Memorial Institute (RPMI)-1640 (HyClone) medium supplemented with 10% fetal bovine serum (FBS, Gibco), 100 U/mL penicillin, and 100 mg/L streptomycin (HyClone) under 37 °C and 5% CO2. Normal and cisplatin-resistant A549 lung cells at the logarithmic growth phase were collected and seeded in 96-well tissue culture plates (5×103 cells/well, Corning), and incubated until adhesion. Cells were treated with cisplatin at indicated concentrations for 48 hours and then 20 µL of MTT (5 mg/mL). Cells were incubated at 37 °C for 4 hours and then treated by 150 µL of dimethyl sulfoxide (DMSO). Microplate Reader Model 550 was used to measure the absorbance value of each well at 490 nm. Every experiment was independently performed for at least three times.
Reverse transcription-quantitative polymerase chain reaction (qRT-PCR)
Total RNA was extracted from normal and cisplatin-resistant A549 lung cells using the TRIzol reagent (Invitrogen), and 500 ng RNA was used for reverse transcription. Stem-loop RT primers were used for miRNAs, and oligo (dT) primer for genes. qRT-PCR was performed on the ABI StepOnePlus™ thermocycler using the SYBR® Green PCR kit. U6 snRNA served as a loading control for miRNAs while beta-actin for genes. Primer sequences are shown in Table 1. All reactions had three replicates. Data were quantified by using the comparative 2−ΔΔCT method.
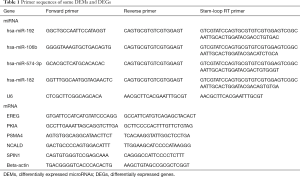
Full table
Statistical analysis
All samples had at least three independent replicates. Data were presented as means ± SD. The SPSS software (version 17.0) was employed for statistical analysis. Statistical significance was examined by t-tests or one-way analysis of variance (ANOVA). P<0.05 (*) or P<0.01 (**) denoted statistically significant difference.
Results
Identification of DEGs
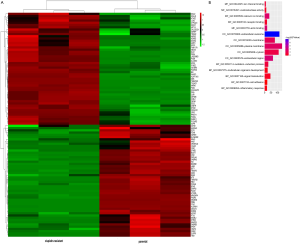
A total of 499 DEGs were selected from E-MEXP-3123. Using the criteria of adj.P.val <0.05 and fold change ≥2.0, 58 upregulated genes and 441 downregulated genes were identified in cisplatin-resistant A549 cells. The top 50 upregulated and downregulated genes were shown in a heatmap (Figure S1A).
GO function and KEGG pathway analyses
To investigate the functions of DEGs, we analyzed GO functions and KEGG pathways. The results were obtained by DAVID. DEGs were significantly involved in some biological processes including signal transduction, oxidation-reduction process, multicellular organism development, cell adhesion and inflammatory response. For molecular functions, DEGs were mainly involved in calcium ion binding, actin binding, receptor binding, oxidoreductase activity and ion channel binding. For cell components, DEGs primarily were enriched in plasma membrane, extracellular exosome, cytosol, membrane and extracellular region (Figure S1B). In addition, KEGG pathway analysis showed that the most significant pathways of DEGs were neuroactive ligand-receptor interaction, endocytosis and hippo signaling pathway (Table 2).
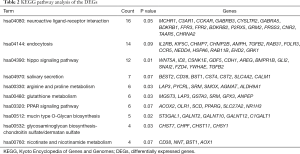
Full table
PPI network and module selection
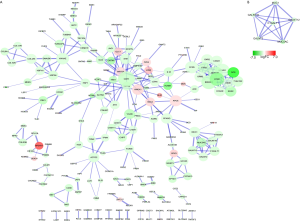
To evaluate the interactions of DEGs, a PPI network with 215 nodes was constructed (Figure S2A). The top five hub genes were CDH1 (cadherin 1), BDKRB2(bradykinin receptor B2), FPR2 (formyl peptide receptor 2), CCR5 (C-C chemokine receptor type 5) and YWHAE (tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein epsilon). In addition, a significant module in PPI network including 6 nodes and 15 edges was selected by MOCODE (Figure S2B).
Identification of DEMs and miRNA-target mRNA network
Twelve DEMs including seven upregulated miRNAs and five downregulated miRNA in cisplatin-resistant A549 cells were identified from GSE43249 (Figure S3A). The top three upregulated miRNAs (miR-194, miR-192 and miR-574-3p) and the top three downregulated miRNAs (miR-106b, miR-146a and miR-182) were uploaded to TargetScan to predict their target genes. The network of miRNAs and their predicted target are shown in Figure S3B.
Experimental validation of the key DEMs in GSE43249 and their potential target genes in E-MEXP-3123
To preliminarily verify the above analyses, we firstly selected the key DEMs in GSE43249 and their potential target genes in E-MEXP-3123. In prediction, the target gene of miR-192 was EREG (epiregulin), miR-106b targeted PKIA (cAMP-dependent protein kinase inhibitor alpha) while miR-182 targeted NCALD (neurocalcin delta) and SPIN1 (spindlin 1), but no prediction was found on miR-194 and miR-146a (Figure 1A).
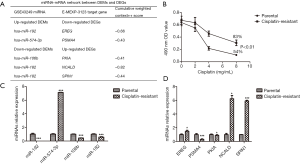
The cisplatin-resistant A549 cells showed higher cell viability than the control group (Figure 1B). After 48 h of cisplatin (8 mg/mL) treatment, the inhibition ratios of cisplatin-resistant group and control group were 83% and 54%, respectively (P<0.01). miRNA and mRNA expression levels were measured by qRT-PCR. As predicted, miR-106b, miR-182, PSMA4 expressions were downregulated in cisplatin-resistant A549 cells, and miR-574-3p, NCALD and SPIN1 expression were upregulated. However, the low expressions of miR-192, PKIA and the high expression of EREG in cisplatin-resistant A549 cells were not consistent with the predicted trends (Figure 1C,D).
Discussion
Drug resistance is a major cause of the clinical failure of chemotherapy and radiotherapy. So far only a few advanced cancers with certain histogenesis can be cured by drugs. Even though these cancers may relapse and obtain drug resistance. Cisplatin is an effective anti-cancer drug and has been used for treating various solid tumors, including testicular, head and neck, ovarian, bladder, colorectal and lung cancers (11). Despite the initial response, many patients develop a cisplatin resistance. Therefore, revealing the specific molecular mechanisms underlying cisplatin resistance is critical for cancer clinical treatment.
Recently, microarray has been an effective tool for studying the expressions of thousands of genes in different physiological and pathological conditions. It enables the discovery of new targets related to drug resistance in tumors. In the present study, 58 significantly upregulated genes and 441 significantly downregulated genes were screened from E-MEXP-3123 which provides the datasets of normal and cisplatin-resistant A549 cells. GO term analysis and KEGG pathway analysis were performed to systematically investigate the regulatory mechanism of DEGs. In GO term analysis, DEGs were significantly involved in some biological processes including signal transduction, oxidation-reduction process, multicellular organism development, cell adhesion and inflammatory response. These results were consistent with the standpoint that cell adhesion mediates drug resistance limiting cancer therapy success (12,13). Low chronic inflammatory response can contribute to drug resistance (14,15). The proteins encoded by both upregulated and downregulated genes were mainly located on the membrane and extracellular exosome. It indicates that cancer drug resistance may be mediated by protein transduction and dissemination (16). Furthermore, pathway analysis showed that the most significant pathways were neuroactive ligand-receptor interaction, endocytosis and hippo signaling pathway.
A PPI network of DEGs was constructed, and the top degree hub genes and a significant module were selected. The top 5 degree hub genes were CDH1, BDKRB2, FPR2, CCR5 and YWHAE. CDH1 has the highest degree in PPI network and is known as Cadherins, which are calcium-dependent cell adhesion proteins. As a famous member of cadherin family, E-cadherin is a ligand for integrin alpha-E/beta-7, and their binding contributes to stabilize epithelial phenotype (17,18). An increasing number of evidence emphasized the close relation between epithelial-mesenchymal transition and drug resistance (19,20). Han et al. found that Cathepsin L increased EMT by downregulating epithelial marker (E-cadherin) and upregulating mesenchymal marker (vimentin), thus mediated cisplatin or paclitaxel resistance in A549 cells (21). The second highest degree hub gene was BDKRB2 which encodes Bradykinin receptor B2 (B2R). It is a G protein-coupled receptor and widely distributed in healthy tissues. It was reported that B2R prevented insulin resistance while B1R is involved in insulin resistance and metabolic syndrome (22). But the relationship between B2R and cancer drug resistance is still unclear. The third highest degree hub gene FPR2 encodes N-FPR2 which is a G-protein coupled receptor and expressed on many human cells including airway epithelium (23). It plays important roles in immune regulation and inflammatory response. A recent study showed that resolvin D1 inhibited TGF-β1-induced EMT in A549 lung cancer cells via binding ALX/FPR2 (24), which indicated that FPR2 may be also involved in drug resistance. CCR5 is located on chromosome 3 and encodes CCR5. It is a protein on the surface of white blood cells that is involved in the immune system as it acts as a receptor for chemokines including CCL3, CCL4 and CCL5, which subsequently transduces a signal by increasing the intracellular calcium ion level (25,26). Recent study showed that XAP, an extract of Marsdenia tenacissima, inhibited A549 cell migration and invasion through down-regulation of CCR5-CCL5 axis, Rho C, and FAK (27). The last hub gene YWHAE encodes a 14-3-3 protein called 14-3-3 protein epsilon. This highly conserved protein implicated in the regulation of diverse biochemical activities related to signal transduction, such as cell division and regulation of insulin sensitivity. Elevated 14-3-3 epsilon level is an independent predictor of chemotherapy-resistance and poor prognosis for patients with advanced extranodal NK/T cell lymphoma with asparaginase-based treatment (28). Taken together, the majority of selected hub genes are involved in A549 lung cancer cells or drug resistance through various biological pathways.
miRNA is a small non-coding RNA molecule with 19–25 nucleotides. It can affect target gene expression to regulate cell apoptosis, proliferation and differentiation. Recently, an increasing number of researches indicate that some miRNAs can target drug-sensitivity genes, leading to cancer drug resistance. In this study, we also analyzed the miRNA expression profiling of cisplatin-resistant cells derived from the A549 lung cell line. Twelve DEMs including seven upregulated miRNAs and five downregulated miRNAs were identified. A network containing the top 6 DEMs and their predicted target genes was constructed. In addition, we selected some hub DEMs in GSE43249 and their target genes in E-MEXP-3123. We cultured the cisplatin-resistant A549 cells and normal A549 cells to verify the above analyses on selected DEMs and their target genes. Our results showed that miR-192 was downregulated and EREG which is miR-192 predicted target was upregulated in cisplatin-resistant A549 cell lines, which was not consistent with our prediction. However, two other studies on cisplatin-resistance in A549 cells showed that miR-192 induced gemcitabine and cisplatin combined chemoresistance by targeting Bcl-2 (29) and cisplatin resistance by targeting Bim (30), respectively. Consistent with our prediction, our study indicated that significantly high expression of miR-574-3p may regulate cisplatin resistance by targeting PSMA4. Ujihira et al. reported that miR-574-3p modulates tamoxifen response in breast cancer (31). Also, our study showed miR-106b and miRNA-182 expressions were downregulated in drug-resistant group. Hu et al. (32) and Ning et al. (33) also showed that miR-106b and miR-182 were closely related to the altered chemosensitivity of human cancer, respectively. Contrary to the prediction, PKIA expression was found to be low in drug-resistant group in our study. This inconsistency between our prediction and results might be due to the prediction error. At present, the role of NCALD or SPIN1 in cisplatin resistance remains unclear. Notably according to our results, NCALD and SPIN1 might be potential targets of miR-182 and play a role in cisplatin-resistant lung cancer, which was completely consistent with the prediction.
The cellular experiment plays an important role in the preliminary verification of further clinical sample experiment. A549 lung cancer cell lines, initiated in 1972 (34), has been widely employed in various studies, such as the molecular pathogenesis of lung cancer and drug function mechanism research. It has been reported that cisplatin can induce both apoptosis and ferroptosis in A549 cells (35). The designed light-triggered nanoparticles called P/C-Micelles exhibit enhanced cisplatin cellular uptake and reduced cisplatin efflux in A549 cells and cisplatin-resistant A549 cells (36). However, it is worth noting that the mechanism of cisplatin resistance in lung cancer cannot be fully understood by only A549 cell lines due to cancer heterogeneity and different living conditions compared with tumor in vivo, more work on some different cell lines and tumor tissues is needed to validate these results in the future.
Data integration and data mining are important tools for understanding the mechanism of cancer drug resistance. In conclusion, in this study, a comprehensive bioinformatical analysis was employed to discover the potential biomarkers for cisplatin resistance. Some significant miRNAs and their predicted DEGs were validated by qRT-PCR. Although these identified genes still need to be verified by more molecular biology experiments and tested in lung cancer tissues, our study might have provided new clues for developing effective strategies against the cisplatin resistance during lung adenocarcinoma treatment.
Acknowledgments
Funding: This work was supported by funds from National Natural Sciences Foundation of China (81502065 and 81672926), China Postdoctoral Science Foundation Funded Project (2016T90613, 2015M580574 and 2016M592146), Natural Sciences Foundation of Shandong Province (ZR2014HQ009), Shandong Postdoctoral Innovation Project (201602037), Qingdao Innovation Applied Basic Research Project (16-5-1-56-JCH) and Qingdao Postdoctoral Research Project (2015167 and 2015157).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.06.15). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This article does not contain any studies with human participants or animals performed by any of the authors. Institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- O'Grady S, Finn SP, Cuffe S, et al. The role of DNA repair pathways in cisplatin resistant lung cancer. Cancer Treat Rev 2014;40:1161-70. [Crossref] [PubMed]
- Tufo G, Jones AW, Wang Z, et al. The protein disulfide isomerases PDIA4 and PDIA6 mediate resistance to cisplatin-induced cell death in lung adenocarcinoma. Cell Death Differ 2014;21:685-95. [Crossref] [PubMed]
- Ishida S, McCormick F, Smith-McCune K, et al. Enhancing tumor-specific uptake of the anticancer drug cisplatin with a copper chelator. Cancer cell 2010;17:574-83. [Crossref] [PubMed]
- Liedert B, Materna V, Schadendorf D, et al. Overexpression of cMOAT (MRP2/ABCC2) is associated with decreased formation of platinum-DNA adducts and decreased G2-arrest in melanoma cells resistant to cisplatin. J Invest Dermatol 2003;121:172-6. [Crossref] [PubMed]
- Yamasaki M, Makino T, Masuzawa T, et al. Role of multidrug resistance protein 2 (MRP2) in chemoresistance and clinical outcome in oesophageal squamous cell carcinoma. Br J Cancer 2011;104:707-13. [Crossref] [PubMed]
- Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447-52. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 2003;4:2. [Crossref] [PubMed]
- Galluzzi L, Senovilla L, Vitale I, et al. Molecular mechanisms of cisplatin resistance. Oncogene 2012;31:1869-83. [Crossref] [PubMed]
- Dickreuter E, Cordes N. The cancer cell adhesion resistome: mechanisms, targeting and translational approaches. Biol Chem 2017;398:721-35. [Crossref] [PubMed]
- Ranji P, Salmani Kesejini T, Saeedikhoo S, et al. Targeting cancer stem cell-specific markers and/or associated signaling pathways for overcoming cancer drug resistance. Tumour Biol 2016;37:13059-75. [Crossref] [PubMed]
- Rasić-Milutinović Z, Perunicić-Peković G, Ristić-Medić D, et al. Insulin resistance and chronic inflammation are associated with muscle wasting in end-stage renal disease patients on hemodialysis. Gen Physiol Biophys 2009;28:184-9. [PubMed]
- Todoric J, Antonucci L, Karin M. Targeting Inflammation in Cancer Prevention and Therapy. Cancer Prev Res (Phila) 2016;9:895-905. [Crossref] [PubMed]
- Qu L, Ding J, Chen C, et al. Exosome-Transmitted lncARSR Promotes Sunitinib Resistance in Renal Cancer by Acting as a Competing Endogenous RNA. Cancer Cell 2016;29:653-68. [Crossref] [PubMed]
- Taraszka KS, Higgins JM, Tan K, et al. Molecular basis for leukocyte integrin alpha(E)beta(7) adhesion to epithelial (E)-cadherin. J Exp Med 2000;191:1555-67. [Crossref] [PubMed]
- Hadley GA, Higgins JM. Integrin alphaEbeta7: molecular features and functional significance in the immune system. Adv Exp Med Biol 2014;819:97-110. [Crossref] [PubMed]
- Elaskalani O, Razak NB, Falasca M, et al. Epithelial-mesenchymal transition as a therapeutic target for overcoming chemoresistance in pancreatic cancer. World J Gastrointest Oncol 2017;9:37-41. [Crossref] [PubMed]
- Brozovic A. The relationship between platinum drug resistance and epithelial-mesenchymal transition. Arch Toxicol 2017;91:605-19. [Crossref] [PubMed]
- Han ML, Zhao YF, Tan CH, et al. Cathepsin L upregulation-induced EMT phenotype is associated with the acquisition of cisplatin or paclitaxel resistance in A549 cells. Acta Pharmacol Sin 2016;37:1606-22. [Crossref] [PubMed]
- Talbot S, Dias JP, El Midaoui A, et al. Beneficial effects of kinin B1 receptor antagonism on plasma fatty acid alterations and obesity in Zucker diabetic fatty rats. Can J Physiol Pharmacol 2016;94:752-7. [Crossref] [PubMed]
- Duvall MG, Levy BD. DHA- and EPA-derived resolvins, protectins, and maresins in airway inflammation. Eur J Pharmacol 2016;785:144-55. [Crossref] [PubMed]
- Lee HJ, Park MK, Lee EJ, et al. Resolvin D1 inhibits TGF-beta1-induced epithelial mesenchymal transition of A549 lung cancer cells via lipoxin A4 receptor/formyl peptide receptor 2 and GPR32. Int J Biochem Cell Biol 2013;45:2801-7. [Crossref] [PubMed]
- Struyf S, Menten P, Lenaerts JP, et al. Diverging binding capacities of natural LD78beta isoforms of macrophage inflammatory protein-1alpha to the CC chemokine receptors 1, 3 and 5 affect their anti-HIV-1 activity and chemotactic potencies for neutrophils and eosinophils. Eur J Immunol 2001;31:2170-8. [Crossref] [PubMed]
- Miyakawa T, Obaru K, Maeda K, et al. Identification of amino acid residues critical for LD78beta, a variant of human macrophage inflammatory protein-1alpha, binding to CCR5 and inhibition of R5 human immunodeficiency virus type 1 replication. J Biol Chem 2002;277:4649-55. [Crossref] [PubMed]
- Lin SS, Li FF, Sun L, et al. Marsdenia tenacissima extract suppresses A549 cell migration through regulation of CCR5-CCL5 axis, Rho C, and phosphorylated FAK. Chin J Nat Med 2016;14:203-9. [PubMed]
- Qiu Y, Zhou Z, Li Z, et al. Pretreatment 14-3-3 epsilon level is predictive for advanced extranodal NK/T cell lymphoma therapeutic response to asparaginase-based chemotherapy. Proteomics Clin Appl 2017;11. [PubMed]
- Cao J, He Y, Liu HQ, et al. MicroRNA 192 regulates chemo-resistance of lung adenocarcinoma for gemcitabine and cisplatin combined therapy by targeting Bcl-2. Int J Clin Exp Med 2015;8:12397-403. [PubMed]
- Zhang F, Li Y, Wu H, et al. MiR-192 confers cisplatin resistance by targeting Bim in lung cancer. Zhongguo Fei Ai Za Zhi 2014;17:384-90. [PubMed]
- Ujihira T, Ikeda K, Suzuki T, et al. MicroRNA-574-3p, identified by microRNA library-based functional screening, modulates tamoxifen response in breast cancer. Sci Rep 2015;5:7641. [Crossref] [PubMed]
- Hu Y, Li K, Asaduzzaman M, et al. MiR-106b~25 cluster regulates multidrug resistance in an ABC transporter-independent manner via downregulation of EP300. Oncol Rep 2016;35:1170-8. [PubMed]
- Ning FL, Wang F, Li ML, et al. MicroRNA-182 modulates chemosensitivity of human non-small cell lung cancer to cisplatin by targeting PDCD4. Diagn Pathol 2014;9:143. [Crossref] [PubMed]
- Lieber M, Smith B, Szakal A, et al. A continuous tumor-cell line from a human lung carcinoma with properties of type II alveolar epithelial cells. Int J Cancer 1976;17:62-70. [Crossref] [PubMed]
- Guo J, Xu B, Han Q, et al. Ferroptosis: A Novel Anti-Tumor Action for Cisplatin. Cancer Res Treat 2017; [Epub ahead of print]. [Crossref] [PubMed]
- Li Y, Deng Y, Tian X, et al. Multipronged Design of Light-Triggered Nanoparticles To Overcome Cisplatin Resistance for Efficient Ablation of Resistant Tumor. ACS Nano 2015;9:9626-37. [Crossref] [PubMed]


