
Epigenetic regulation in bladder cancer: development of new prognostic targets and therapeutic implications
Introduction
Bladder cancer is one of the leading cancers of the urinary tract. It is often diagnosed at advanced stages of the disease and high rates of recurrence are the most difficult obstacle for patient treatment (1). To explore early diagnostic and possible therapeutic targets for bladder cancer, understanding the regulatory system of gene expression is one of the most significant steps. Among many transcriptional regulatory mechanisms, modulation of epigenetic modifications has been studied most extensively over the last decade. Epigenetic modifications to DNA or its associated proteins result in changes in gene expression without altering DNA sequences (2). These characteristics make regulation of epigenetic mechanisms more flexible and important therapeutic targets for cancer treatments. Epigenetic regulatory modification includes DNA methylation alterations to gene promoter regions, histone modifications, and microRNA (miRNA) expression changes. DNA methylation is an enzymatic process involving covalent modifications of DNA with methyl groups using S-adenosyl methionine (SAM) as the methyl group donor and addition of this moiety to the 5-carbon atom of the cytosine base. DNA methylation typically acts to repress gene transcription. In plants and other organisms, DNA methylation is generally found in three different sequence contexts: CG (or CpG), CHG, or CHH (where H corresponds to A, T, or C). In mammals, however, DNA methylation almost exclusively occurs at CpG dinucleotide sites, with the cytosine on both strands usually being methylated. The DNA methylation process is known to be associated with development, aging, and carcinogenesis (3). In tumorigenesis, hypermethylation of tumor suppressor genes and hypomethylation of oncogenes have been characterized. Histone modification includes acetylation, methylation, carbonylation, ubiquitination, SUMOylation, and phosphorylation of histone side chains (4). The functional unit of chromatin, the nucleosome, consists of four histone proteins identified as H2A, H2B, H3, and H4. Each of these proteins exhibit ‘tail’ extensions and these can be modified by a number of histone modifying enzymes. The addition of specific chemical groups to histone tails usually alters the binding affinity of the tails to DNA and results in changes in the conformation of the chromatin structure to regulate RNA transcription of genes. Histone acetylation usually indicates active gene transcription, whereas histone methylation is associated with both gene repression and activation, depending on the specific target histone residues. In this review, genome-wide histone modification and expression of key histone modifying enzymes associated with bladder cancer will be discussed. A miRNA is a small non-coding RNA molecule found in various species including plants, animals, and some viruses. They are approximately 18 to 25-nucleotide-long and are synthesized and processed in the nucleus. The function of miRNAs involves RNA silencing and post-transcriptional regulation of gene expression (5). It has been estimated that at least 30% of human genes may be regulated by interactions with miRNAs. It is established that each mRNA can interact with multiple miRNAs and that each miRNA may regulate multiple mRNAs. miRNAs play an important role in tumorigenesis by activating oncogenes or restraining tumor suppressor genes (6). We will explore and discuss epigenetic regulation of tumor-related genes in bladder cancer, and possible therapeutic and diagnostic targets for bladder cancer therapy. The diagnosis approach for bladder cancer is typically invasive causing discomfort to patients and only provides a generalized outcome for patients. The need for non-invasive markers and demand for improved screening techniques are urgent. We will also discuss a number of epigenetic biomarkers that have emerged recently as candidates for more effective diagnosis for bladder cancer.
DNA methylation in bladder cancer
Cancer initiation and progression are often accompanied by profound changes in DNA methylation which are marked by genome-wide hypomethylation and site-specific CpG island promoter hypermethylation (7). Hypomethylation occurs mainly in repetitive DNA sequences such as tandem and interspersed repeats and in segmental duplications. This leads to activation of cancer-causing genes, with global DNA hypomethylation often being associated with metastatic disease. Hypermethylation is associated with CpG islands and mediates silencing of tumor suppressor genes commonly associated with cancer development. Activity of DNA methyltransferases (DNMTs) generates a pattern of methylated CpG di-nucleotides at the 5' region of the promoter of the gene involved.
In bladder cancer, a large number of genes have been shown to undergo promoter hypermethylation. These loci include GSTP1, TIG1, TIMP-2, p14ARF, RUNX3, APC, SFRP2, RASSF1A, MGMT, FHIT, CDH13, and RARB (8-17) (Table 1). In particular, the RUNX3 gene was implicated as a tumor suppressor and exhibited significant elevations of methylation levels in bladder cancer upon analysis of 124 tumor tissue samples (14). A prognostic indicator in patients with non-muscle invasive bladder cancer (NMIBC) was found by Yoon et al. (20). Their analysis of 136 human bladder specimens (8 normal controls and 128 NMIBCs) showed clinical relevance of the radial spoke head 9 homolog (RSPH9) gene methylation status as an independent predictor of disease recurrence and progression. Another report suggested that hypermethylation of PCDH10 and PCDH17 was related to bladder cancer development and cancer-specific survival (21,22). A recent paper reported that the combination of PCDH17 and POU4F2 methylation in urine sediment DNA yielded high sensitivity and specificity (90% and 93.96%, respectively) in detecting bladder cancer (19). Renard et al. showed that identification of TWIST1 and NID2 methylation can be employed for detecting bladder cancer with high sensitivity and specificity (93% and 90%, respectively) in urine samples (18). Targeted epigenetic analysis using tumor biopsies (n=105) and urine samples (n=113) revealed 95 oncogenic mutations and 189 hypermethylation events. They revealed that three methylation markers, including APC, RASSF1A, and SFRP2, together with genetic mutation of FGFR3 provided sensitivity of 90% for tumor specimens and 62% for urine samples, with 100% specificity respectively (23). Genomic DNA hypomethylation is considered to be a biomarker for bladder cancer with elevated long interspersed element-1 (LINE-1) hypomethylation (24). LINE-1 hypomethylation was found to be associated with oxidative stress which may contribute to the promotion of urothelial cell carcinogenesis (25).
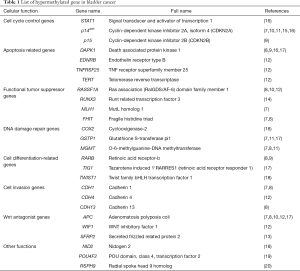
Full table
Although some obstacles arising from the employment of bisulfite treatment need to be surmounted, including DNA isolation efficiency, leukocyte DNA contamination, and loss of DNA templates, these DNA hypermethylation markers may improve the understanding of tumorigenesis and the prognostic value of DNA-based diagnosis in noninvasive detection of bladder cancer. Several chemical agents used for bladder cancer treatment exhibit DNA methyltransferase inhibitor activity. 5-Fluoro-2-deoxycytidine is such an inhibitor and was used together with tetrahydrouridine, a cytosine deamination inhibitor, and they synergistically inhibited bladder cancer growth (26). The combination of DNA methyltransferase inhibitor and tetrahydrouridine for bladder cancer treatment is now in phase II clinical trials. When the anti-metabolite gemcitabine was tested against bladder cancer cell lines, the DNA methylation status of several epigenetically silenced genes, including GSTP1, IGFBP3, and RASSF1A, was changed (27)
MicroRNA regulation in bladder cancer growth
Since their discovery in the early 21st century, miRNAs have been intensively studied in various fields, including development, differentiation, metabolism, and cancer progression. miRNAs, a class of noncoding RNAs, is approximately 18 to 25-nucleotide-long and can be synthesized and processed in the nucleus. They are encoded within intergenic regions or within introns or exons of protein-coding genes of the genome. The miRNA sequence is initially transcribed to a several-kb-long primary transcript, which is subsequently processed by diverse enzymes, including exportin-5 and its catalytic partner. After translocation to the cytosol, miRNAs are enzymatically processed by Dicer to approximately 22-nucleotide-long mature miRNAs. These can bind matching mRNAs and regulate their stability (28). In tumor tissues, the expression of some miRNAs has been reported to be related to cell proliferation, differentiation, and apoptotic processes. The overexpression of these miRNAs can diminish expression of tumor suppressor genes and is referred to as oncogenic miRNAs.
Ten human micro-RNAs, including miR-223, miR-26b, miR-221, miR-103-1, miR-185, miR-23b, miR-203, miR-17-5p, miR-23a, and miR-205, were reported to be significantly up-regulated in bladder cancer specimens. In the same study, only miR-26b was shown to exhibit decreased abundance (29). A recent study suggested that miR-143 expression levels were suppressed in human bladder cancer tissues and cells, and such diminished expression levels were negatively correlated with bladder cancer clinical staging. The authors proposed that the functional target of miR-143 in bladder cancer was insulin-like growth factor-1 receptor (IGF-1R), and this was correlated with patient survival (30). Another study reported that miR-24-3p was highly expressed in bladder cancer tissues and functioned to regulate DEDD, a member of the death effector domain-containing protein family. DEDD was seen to be expressed at low levels in bladder cancer. Western blotting and RT-PCR experiments revealed that miR-24-3p inhibited apoptosis and promoted the growth and proliferation of bladder cancer cells (31). The expression level of miR-5195-3p was also down-regulated in bladder cancer samples, and the target of miR-5195-3p was identified as the Krüppel-like transcription factor-5 (KLF5), a proven proto-oncogene (32). After bioinformatics screening for epigenetic regulators of Survivin, an important enzyme in bladder cancer development, miR-138-5p was confirmed to act as a 3'-UTR-targeting modulator of Survivin (33). The functions of miR-138-5p in bladder cancer growth and invasive characteristics were confirmed in a xenograft mouse model. miR-106a also regulated cell cycle and proliferation processes in bladder cancer (34).
Despite their clinical significance in tumorigenesis, little is known about the function of miRNAs in cancer metastasis. Noteworthy, various studies regarding miRNA functions and involvement in bladder cancer metastasis were published over several years. Immunohistological screening of miRNAs by tissue microarray revealed that expression of miR-3658 was upregulated in bladder cancer tissues, and is associated with lymph node invasion, distant metastasis, and histological grading (35). Chemokine receptor type 7 (CCR7) is known to be correlated with lymphatic metastasis and poor clinical prognosis in bladder cancer. MiR-199a-5p was found to target the 3'-UTR of CCR7 and this correlation was further strongly linked to the miRNA/chemokine axis that regulates bladder cancer cell metastasis (36). miR-3713 also regulates MMP9 in bladder cancer and significantly lower levels have been found in bladder cancer tissues (37). Data mining reports using public databases also suggested a correlation between miRNAs and several EMT genes. Two miRNAs, miR-214-3p and miR-145-5p were reported to target and modulate MMP-9 and NGAL genes (38). miR-451 was found to regulate bladder cancer cell migration and invasion by directly targeting c-Myc, and apoptosis promoted by miR-451 may participate in this process (39). When miR-106a was expressed in bladder cancer cell lines, migration and invasive capacity of these cell lines decreased. miR-101 was found to be less expressed in bladder cancer tissues and bladder epithelial cell lines (40,41). TargetScan analysis and a luciferase assay confirmed that miR-101 negatively regulates c-FOS levels. Bladder cancer cell proliferation and invasion were inhibited by miR-101. The above mentioned miRNAs dysregulated in bladder cancer were listed in Table 2.

Full table
There are some therapeutic candidate inhibitor miRNAs potentially useful for bladder cancer treatment. Gambogic acid (GA), an inhibitor of zeste homolog 2 (EZH2), was shown to induce bladder cancer cell apoptosis, mediated through miR-101 (42). This result established GA as a possible combination therapeutic drug for bladder cancer treatment. Another natural product, honokiol, isolated from the Magnolia plant, inhibited bladder cancer cell proliferation, survival, cancer stemness, migration, and invasion through down-regulation of EZH2 expression. This effect was associated with miR-143, a tumor suppressor micro RNA (43).
Histone modification in bladder cancer
Histones are small, highly-conserved proteins and comprise the key protein components of chromatin condensation, which packs DNA into chromosome domains. Histone modification includes methylation, acetylation, phosphorylation, ubiquitination, and SUMOylation of particular amino acids, usually at N-terminal tails. They are known to play crucial roles in regulating many fundamental biological processes, including gene expression, DNA replication, and DNA damage repair (44). Acetylation of histone residues usually represents active chromatin regions, whereas methylation may be associated with both gene activation and repression, depending on the specific lysine residue position (45). In bladder cancer, a growing number of papers are being published regarding the relationship between histone modifications and cancer progression (Table 3). Genetic profiling recently revealed that 89% of bladder cancers exhibit altered histone modification pathways, and 64% of cancers harbor alterations in the SWI/SNF complex, which is responsible for chromatin remodeling so as to activate or inactivate gene transcription (62).
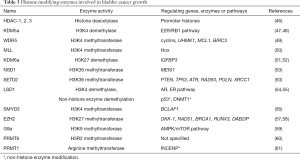
Full table
Histone acetylation in bladder cancer
A recent study on histone acetylation reported that global H3 acetylation (H3Ac) levels decreased in bladder cancer patients, whereas H3K18Ac and H4Ac levels were similar between normal and cancer group (63). Another report examining tissue microarray involved 174 bladder cancer patients and investigated expression levels of histone deacetylases (HDACs) 1, 2 and 3. High-grade noninvasive papillary bladder tumors were associated with elevated expression levels of HDAC-1 and HDAC-2 (46). In particular, high grade tumors, in combination with high expression of HDAC-1, exhibited poorer prognoses than other tumors did. These results indicate that inhibition of HDAC has potential in bladder cancer therapy. Romidepsin, a histone deacetylase inhibitor effective in T-cell lymphoma therapy, was reported to have suppressive effects on bladder cancer growth in combination with tetrahydrouridine, and currently, is in phase I trials for bladder cancer treatment (64). Combined treatment with the histone deacetylase inhibitor OBP-801 and celecoxib demonstrated bladder cancer cell growth inhibition and promoted apoptosis (65). The authors showed that this combined treatment was dependent on the expression of death receptor 5 (DR5), and knockdown of DR5 significantly suppressed cell apoptosis with this combined treatment regime. Another histone deacetylase inhibitor, AR-42, has shown promise in combination with cisplatin in treating bladder cancer in a mouse model and in in vitro stem cell culture (66). The histone deacetylase inhibitor trichostatin A re-sensitized the otherwise gemcitabine-resistant urothelial carcinoma cells via suppression of triglyceride (TG)-interacting factor and down-regulating Akt signaling (67). Transient receptor potential cation channel, subfamily M, member 2 (TRPM2) protein is reportedly controlled by histone deacetylase inhibition, with its promoter acetylation status (68). TRPM2 up-regulation by an HDAC inhibitor promoted apoptosis in bladder cancer cell lines, and TRPM2 promoter acetylation changes were monitored by chromatin immunoprecipitation (ChIP) experiments. A proteomic approach revealed that histone deacetylase inhibitors (including romidepsin, trichostatin A, vorinostat) induced bladder cancer cell death, which is associated with chromatin modification and modulation of protein expression of some 6,003 proteins (69).
Histone lysine methylation in bladder cancer
In 2011, a report showed that global histone H4K20 tri-methylation is related to cancer-specific survival in muscle-invasive bladder cancer patients (70). Furthermore, numerous specific histone modification enzymes are reported to be involved in bladder cancer growth and proliferation. Depletion of the H3K4 demethylase KDM5a resulted in cell growth retardation through co-regulation of the E2F/RB1 pathway in bladder cancer (47,48). One of the core components of the MLL/SET1 complex is WDR5, which exerts methyltransferase activity. WDR5 was found to be highly expressed in bladder cancer tissues, and overexpression of WDR5 promoted bladder cancer cell growth in vitro and in vivo (49). Whole-exome sequencing of 37 bladder cancer patient tissue samples revealed that MLL, EP400, PRDM2, ANK3, and CHD5 were differentially altered in recurrent bladder tumors (50). The authors also showed that MLL-mutated bladder cancer cells using CRISPR/Cas9 genetic modification demonstrated increased drug-resistance to epirubicin (a chemotherapeutic drug employed for patients with bladder cancer). Additionally, alterations in the MLL gene were involved in bladder cancer relapse. Another massive exome sequencing using 54 bladder cancer samples revealed that KDM6A and BRCA1-associated protein-1 (BAP-1) mutations frequently co-occurred in tumors. Depletion of KDM6a in bladder cancer cell lines enhanced anchorage independent growth and cell migration, but not monolayer growth (51,52). These data showed that KMD6A plays a role in tumor growth and suppression of cell migration. When a NF-kB inhibitor, dimethylaminoparthenolide (DMAPT) was tested for potential effects on bladder cancer cell proliferation, H3K36 trimethylase NSD1 (KMT3B) and SETD2 (KMT3A) levels increased and cell death was induced (53). Lysine-specific demethylase 1 (LSD1) was found to be highly expressed in bladder cancer stem cells, and knock down of LSD1 in bladder cancer cell resulted in significant suppression of cancer proliferation (54). A study involving Drosophila unveiled the function of LSD1 in Pumilio (PUM1), a post-transcriptional repressor that plays important roles in bladder cancer development (55). Up-regulation of the histone methyl transferase SMYD3 promoted bladder cancer progression by targeting BCL2-associated transcription factor 1 (BCLAF1) promoter (56). The authors demonstrated that SMYD3 physically interacted with the BCLAF1 promoter and upregulated its expression by accumulating di- and tri-methylation of H3K4. Upregulated BCLAF1 increased autophagy and bladder cancer cell growth. Enhancer of EZH2 belongs to the Polycomb repressive complex 2, acting as its catalytic subunit, and which exerts gene silencing effects through the trimethylation of H3K27. EZH2 was reported to be overexpressed in bladder cancer and involved in development and progression of bladder cancer (57,58). G9a H3K9 methyltransferase is known as an oncogene in bladder cancer, and inhibition of G9a induced autophagic cell death via the AMPK/mTOR pathway in bladder cancer cell lines (59).
A small-molecule G9a inhibitor, BIX-01294, was reported to inhibit tumor cell growth and also induce apoptosis of bladder cancer cells through the ER stress pathway, resulting in PMAIP1 up-regulation and MCL1 down-regulation (71). Emodin, a natural product, which has the ability to inhibit bladder cancer cell line growth, was found to enhance H3K27 trimethylation. Today, various inhibitors targeting HKMTs and HKDMs are being increasingly developed for cancer treatment (72).
Histone arginine methylation in bladder cancer
Histone arginine methylation in bladder cancer has, to date, been studied rather less than has histone lysine methylation. PRMT6 is a type I protein arginine methyltransferase, and is reported to act on H3R2, which has been identified as a gene repressive epigenetic mark. PRMT6 was reported to be overexpressed in bladder cancer (60). Protein arginine methyltransferase PRMT1 was observed to be overexpressed in bladder cancer (61) and promotes mitosis of cancer cells through inner centromere protein (INCENP) arginine methylation (61). Although less well-studied than lysine methylation, arginine methylation may also contribute to the initiation, progression and development of bladder cancer and likely represents a promising target for therapeutic drug screening.
Clinical approaches targeting epigenetic regulators
Several molecules targeting epigenetic alterations have been developed for anticancer therapy (Table 4). Among them, some small molecule inhibitors have been approved by the U.S. Food and Drug Administration, and were proven therapeutically effective in various cancers (73). DNMT inhibitor 5-aza-2'-deoxycytidine (5-aza-CR; Vidaza®; decitabine) was approved for myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML); in addition, it completed phase 2 trials for treatment of bladder cancer (74), and it is in phase 1 trials in combination with tetrahydrouridine for bladder cancer treatment (64). Inhibition of DNA methylation with cytosine deamination (5-fluoro-2-deoxycytidine with tetrahydrouridine) have finished phase 2 trials in bladder cancer (26). Romidepsin (depsipeptide or Istodax), a HDAC inhibitor approved for cutaneous T-cell lymphoma (CTCL) in 2009, has been in phase 2 trials for treatment of many cancer types, including bladder cancer (75). Another HDAC inhibitor, vorinostat (SAHA, Zolinza®, MK-0683) was approved for CTCL in 2006, passed phase 2 trials as a monotherapy for bladder cancer and completed phase 1 trials as a combination therapy with docetaxel (76). Valproic acid, a short-chain fatty acid, which exhibits histone deacetylase inhibitory activity, is currently in phase 1–2 clinical trials either alone or in combination with other drugs, such as azacitidine, retinoic acid, temozolomide, and karenitecin, for treatment of some solid tumors (77). Tazemetostat (an EZH2 histone N-methyl transferase inhibitor) is being studied in ongoing clinical trials. For the diagnostic use of epigenetic regulators for treating bladder cancer patients, the following article may be a good reference. Targeted exome sequencing of 50 bladder cancer patients revealed that many epigenetic regulator genes were mutated in cancer specimens, and this results can be used for targeted therapy for bladder cancer patients at very high risk of metastasis and death (78). Such high-throughput technologies and clear understanding of epigenetic regulators could open the way for the use of personalized targeted therapies for bladder cancer treatment.

Full table
Conclusions
Invasive bladder cancer is characterized by high rates of mutation in many cancer related genes. Identifying epigenetic changes and their multiple and significant roles in bladder cancer development represent highly useful targets for patient diagnosis and drug screening. Epigenetic profiling also can allow identification of patients’ risk of tumor invasiveness using less intrusive urine tests. Epigenetic modifiers such as histone deacetylase and methyl transferases have come into focus as novel drug targets for bladder cancer therapy. The importance of epigenetics in bladder cancer has now been fully recognized and the research in this arena has matured evolved over the years. The use of high throughput methods has made it possible to scrutinize epigenetic processes (and their effects) at the global level than those involving individual genes. Next-generation sequencing techniques combined with chromatin immune-precipitation (Chip-seq) have enabled studies of the histone modification status of bladder cancer patients at a global level. In addition, next-generation sequencing also allowed studies of DNA methylation status at the single nucleotide level. Employing these epigenetic data will help profoundly in designing more potent treatment strategies in bladder cancer. Moreover, the intrinsic reversibility nature of epigenetic modifications will afford exciting new opportunities for the development and establishment of improved, novel strategies for bladder cancer prevention, detection and therapeutic intervention. An important note is that, due to the crucial functions of epigenetic control and regulation in normal (as well as abnormal) physiology, inhibition could potentially exert toxic effects on non-diseased organs. Hence, toxicological and combination therapy studies should be performed carefully in the development of new drugs targeting epigenetic pathways.
Acknowledgments
Funding: This research was supported by the Basic Science Research Program through the National Research Foundation (NRF) of Korea and funded by the Ministry of Science, ICT & Future Planning (NRF-2015R1C1A2A01051546) and an Incheon National University (International Cooperative) Research Grant in 2016.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Ja Hyeon Ku, Kunyoo Shin, Minyong Kang) for the series “Bladder Cancer” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.05.41). The series “Bladder Cancer” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Raghavan D, Shipley WU, Garnick MB, et al. Biology and management of bladder cancer. N Engl J Med 1990;322:1129-38. [Crossref] [PubMed]
- Petronis A. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature 2010;465:721-7. [Crossref] [PubMed]
- Schübeler D. Function and information content of DNA methylation. Nature 2015;517:321-6. [Crossref] [PubMed]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature 2000;403:41-5. [Crossref] [PubMed]
- Ambros V. The functions of animal microRNAs. Nature 2004;431:350-5. [Crossref] [PubMed]
- Adams BD, Parsons C, Walker L, et al. Targeting noncoding RNAs in disease. J Clin Invest 2017;127:761-71. [Crossref] [PubMed]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002;3:415-28. [PubMed]
- Maruyama R, Toyooka S, Toyooka KO, et al. Aberrant Promoter Methylation Profile of Bladder Cancer and Its Relationship to Clinicopathological Features. Cancer Res 2001;61:8659-63. [PubMed]
- Chan MW, Chan LW, Tang NL, et al. Hypermethylation of Multiple Genes in Tumor Tissues and Voided Urine in Urinary Bladder Cancer Patients. Clin Cancer Res 2002;8:464-70. [PubMed]
- Dulaimi E, Uzzo RG, Greenberg RE, et al. Detection of Bladder Cancer in Urine by a Tumor Suppressor Gene Hypermethylation Panel. Clin Cancer Res 2004;10:1887-93. [Crossref] [PubMed]
- Hoque MO, Begum S, Topaloglu O, et al. Quantitation of Promoter Methylation of Multiple Genes in Urine DNA and Bladder Cancer Detection. J Natl Cancer Inst 2006;98:996-1004. [Crossref] [PubMed]
- Yates DR, Rehman I, Abbod MF, et al. Promoter Hypermethylation Identifies Progression Risk in Bladder Cancer. Clin Cancer Res 2007;13:2046-53. [Crossref] [PubMed]
- Marsit CJ, Karagas MR, Andrew A, et al. Inactivation of SFRP Genes and TP53 Alteration Act Jointly as Markers of Invasive Bladder Cancer. Cancer Res 2005;65:7081-5. [Crossref] [PubMed]
- Kim EJ, Kim YJ, Jeong P, et al. Methylation of the RUNX3 Promoter as a Potential Prognostic Marker for Bladder Tumor. J Urol 2008;180:1141-5. [Crossref] [PubMed]
- Domínguez G, Carballido J, Silva J, et al. p14ARF promoter hypermethylation in plasma DNA as an indicator of disease recurrence in bladder cancer patients. Clin Cancer Res 2002;8:980-5. [PubMed]
- Friedrich MG, Chandrasoma S, Siegmund KD, et al. Prognostic relevance of methylation markers in patients with non-muscle invasive bladder carcinoma. Eur J Cancer 2005;41:2769-78. [Crossref] [PubMed]
- Ellinger J, El Kassem N, Heukamp LC, et al. Hypermethylation of Cell-Free Serum DNA Indicates Worse Outcome in Patients With Bladder Cancer. J Urol 2008;179:346-52. [Crossref] [PubMed]
- Renard I, Joniau S, van Cleynenbreugel B, et al. Identification and validation of the methylated TWIST1 and NID2 genes through real-time methylation-specific polymerase chain reaction assays for the noninvasive detection of primary bladder cancer in urine samples. Eur Urol 2010;58:96-104. [Crossref] [PubMed]
- Wang Y, Yu Y, Ye R, et al. An epigenetic biomarker combination of PCDH17 and POU4F2 detects bladder cancer accurately by methylation analyses of urine sediment DNA in Han Chinese. Oncotarget 2016;7:2754-64. [PubMed]
- Yoon HY, Kim YJ, Kim JS, et al. RSPH9 methylation pattern as a prognostic indicator in patients with non-muscle invasive bladder cancer. Oncol Rep 2016;35:1195-203. [PubMed]
- Guil S, Esteller M. DNA methylomes, histone codes and miRNAs: tying it all together. Int J Biochem Cell Biol 2009;41:87-95. [Crossref] [PubMed]
- Lin YL, Li ZG, He ZK, et al. Clinical and prognostic significance of protocadherin-10 (PCDH10) promoter methylation in bladder cancer. J Int Med Res 2012;40:2117-23. [Crossref] [PubMed]
- Serizawa RR, Ralfkiaer U, Steven K, et al. Integrated genetic and epigenetic analysis of bladder cancer reveals an additive diagnostic value of FGFR3 mutations and hypermethylation events. Int J Cancer 2011;129:78-87. [Crossref] [PubMed]
- Patchsung M, Boonla C, Amnattrakul P, et al. Long Interspersed Nuclear Element-1 Hypomethylation and Oxidative Stress: Correlation and Bladder Cancer Diagnostic Potential. PLoS ONE 2012;7:e37009 [Crossref] [PubMed]
- Wongpaiboonwattana W, Tosukhowong P, Dissayabutra T, et al. Oxidative stress induces hypomethylation of LINE-1 and hypermethylation of the RUNX3 promoter in a bladder cancer cell line. Asian Pac J Cancer Prev 2013;14:3773-8. [Crossref] [PubMed]
- Suzuki T, Tanaka R, Hamada S, et al. Design, synthesis, inhibitory activity, and binding mode study of novel DNA methyltransferase 1 inhibitors. Bioorg Med Chem Lett 2010;20:1124-7. [Crossref] [PubMed]
- Gray SG, Baird AM, O'Kelly F, et al. Gemcitabine reactivates epigenetically silenced genes and functions as a DNA methyltransferase inhibitor. Int J Mol Med 2012;30:1505-11. [PubMed]
- Piletič K, Kunej T. MicroRNA epigenetic signatures in human disease. Arch Toxicol 2016;90:2405-19. [Crossref] [PubMed]
- Gottardo F, Liu CG, Ferracin M, et al. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol 2007;25:387-92. [Crossref] [PubMed]
- Wang H, Li Q, Niu X, et al. miR-143 inhibits bladder cancer cell proliferation and enhances their sensitivity to gemcitabine by repressing IGF-1R signaling. Oncol Lett 2017;13:435-40. [PubMed]
- Yu G, Jia Z, Dou Z. miR-24-3p regulates bladder cancer cell proliferation, migration, invasion and autophagy by targeting DEDD. Oncol Rep 2017;37:1123-31. [PubMed]
- Jiang Z, Zhang Y, Cao R, et al. MiR-5195-3p inhibits proliferation and invasion of human bladder cancer cells by directly targeting oncogene KLF5. Oncol Res 2017; [Epub ahead of print]. [Crossref] [PubMed]
- Yang R, Liu M, Liang H, et al. miR-138-5p contributes to cell proliferation and invasion by targeting Survivin in bladder cancer cells. Mol Cancer 2016;15:82. [Crossref] [PubMed]
- Shin SS, Park SS, Hwang B, et al. MicroRNA-106a suppresses proliferation, migration, and invasion of bladder cancer cells by modulating MAPK signaling, cell cycle regulators, and Ets-1-mediated MMP-2 expression. Oncol Rep 2016;36:2421-9. [PubMed]
- Chen YJ, Wang HF, Liang M, et al. Upregulation of miR-3658 in bladder cancer and tumor progression. Genet Mol Res 2016;15: [Crossref] [PubMed]
- Zhou M, Wang S, Hu L, et al. miR-199a-5p suppresses human bladder cancer cell metastasis by targeting CCR7. BMC Urol 2016;16:64. [Crossref] [PubMed]
- Wu WB, Wang W, Du YH, et al. MicroRNA-3713 regulates bladder cell invasion via MMP9. Sci Rep 2016;6:32374. [Crossref] [PubMed]
- Falzone L, Candido S, Salemi R, et al. Computational identification of microRNAs associated to both epithelial to mesenchymal transition and NGAL/MMP-9 pathways in bladder cancer. Oncotarget 2016;7:72758-66. [PubMed]
- Wang J, Zhao X, Shi J, et al. miR-451 suppresses bladder cancer cell migration and invasion via directly targeting c-Myc. Oncol Rep 2016;36:2049-58. [PubMed]
- Hu Z, Lin Y, Chen H, et al. MicroRNA-101 suppresses motility of bladder cancer cells by targeting c-Met. Biochem Biophys Res Commun 2013;435:82-7. [Crossref] [PubMed]
- Long Y, Wu Z, Yang X, et al. MicroRNA-101 inhibits the proliferation and invasion of bladder cancer cells via targeting c-FOS. Mol Med Rep 2016;14:2651-6. [PubMed]
- Wang Y, Xiang W, Wang M, et al. Methyl jasmonate sensitizes human bladder cancer cells to gambogic acid-induced apoptosis through down-regulation of EZH2 expression by miR-101. Br J Pharmacol 2014;171:618-35. [Crossref] [PubMed]
- Zhang Q, Zhao W, Ye C, et al. Honokiol inhibits bladder tumor growth by suppressing EZH2/miR-143 axis. Oncotarget 2015;6:37335-48. [PubMed]
- Jenuwein T, Allis CD. Translating the Histone Code. Science 2001;293:1074-80. [Crossref] [PubMed]
- Kouzarides T. Chromatin Modifications and Their Function. Cell 2007;128:693-705. [Crossref] [PubMed]
- Poyet C, Jentsch B, Hermanns T, et al. Expression of histone deacetylases 1, 2 and 3 in urothelial bladder cancer. BMC Clin Pathol 2014;14:10. [Crossref] [PubMed]
- Hayami S, Yoshimatsu M, Veerakumarasivam A, et al. Overexpression of the JmjC histone demethylase KDM5B in human carcinogenesis: involvement in the proliferation of cancer cells through the E2F/RB pathway. Mol Cancer 2010;9:59. [Crossref] [PubMed]
- Li X, Su Y, Pan J, et al. Connexin 26 is Down-Regulated by KDM5B in the Progression of Bladder Cancer. Int J Mol Sci 2013;14:7866. [PubMed]
- Chen X, Xie W, Gu P, et al. Upregulated WDR5 promotes proliferation, self-renewal and chemoresistance in bladder cancer via mediating H3K4 trimethylation. Sci Rep 2015;5:8293. [Crossref] [PubMed]
- Wu S, Yang Z, Ye R, et al. Novel variants in MLL confer to bladder cancer recurrence identified by whole-exome sequencing. Oncotarget 2016;7:2629-45. [PubMed]
- Yap KL, Kiyotani K, Tamura K, et al. Whole-exome sequencing of muscle-invasive bladder cancer identifies recurrent mutations of UNC5C and prognostic importance of DNA repair gene mutations on survival. Clin Cancer Res 2014;20:6605-17. [Crossref] [PubMed]
- Ler LD, Ghosh S, Chai X, et al. Loss of tumor suppressor KDM6A amplifies PRC2-regulated transcriptional repression in bladder cancer and can be targeted through inhibition of EZH2. Sci Transl Med 2017;9: [Crossref] [PubMed]
- Nakshatri H, Appaiah HN, Anjanappa M, et al. NF-[kappa]B-dependent and -independent epigenetic modulation using the novel anti-cancer agent DMAPT. Cell Death Dis 2015;6:e1608 [Crossref] [PubMed]
- Lan W, Zhang D, Jiang J. The roles of LSD1-mediated epigenetic modifications in maintaining the pluripotency of bladder cancer stem cells. Med Hypotheses 2013;81:823-5. [Crossref] [PubMed]
- Miles WO, Lepesant JM, Bourdeaux J, et al. The LSD1 Family of Histone Demethylases and the Pumilio Posttranscriptional Repressor Function in a Complex Regulatory Feedback Loop. Mol Cell Biol 2015;35:4199-211. [Crossref] [PubMed]
- Shen B, Tan M, Mu X, et al. Upregulated SMYD3 promotes bladder cancer progression by targeting BCLAF1 and activating autophagy. Tumour Biol 2016;37:7371-81. [Crossref] [PubMed]
- Yamaguchi H, Hung MC. Regulation and Role of EZH2 in Cancer. Cancer Res Treat 2014;46:209-22. [Crossref] [PubMed]
- Martínez-Fernández M, Rubio C, Segovia C, et al. EZH2 in Bladder Cancer, a Promising Therapeutic Target. Int J Mol Sci 2015;16:27107-32. [PubMed]
- Li F, Zeng J, Gao Y, et al. G9a Inhibition Induces Autophagic Cell Death via AMPK/mTOR Pathway in Bladder Transitional Cell Carcinoma. PLoS ONE 2015;10:e0138390 [Crossref] [PubMed]
- Wu H, Zheng W, Eram MS, et al. Structural basis of arginine asymmetrical dimethylation by PRMT6. Biochem J 2016;473:3049-63. [Crossref] [PubMed]
- Deng X, Von Keudell G, Suzuki T, et al. PRMT1 promotes mitosis of cancer cells through arginine methylation of INCENP. Oncotarget 2015;6:35173-82. [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014;507:315-22. [Crossref] [PubMed]
- Ellinger J, Schneider AC, Bachmann A, et al. Evaluation of Global Histone Acetylation Levels in Bladder Cancer Patients. Anticancer Res 2016;36:3961-4. [PubMed]
- Bertino EM, Otterson GA. Romidepsin: a novel histone deacetylase inhibitor for cancer. Expert Opin Investig Drugs 2011;20:1151-8. [Crossref] [PubMed]
- Toriyama S, Horinaka M, Yasuda S, et al. A Histone Deacetylase Inhibitor, OBP-801, and Celecoxib Synergistically Inhibit the Cell Growth with Apoptosis via a DR5-Dependent Pathway in Bladder Cancer Cells. Mol Cancer Ther 2016;15:2066-75. [Crossref] [PubMed]
- Li DR, Zhang H, Peek E, et al. Synergy of Histone-Deacetylase Inhibitor AR-42 with Cisplatin in Bladder Cancer. J Urol 2015;194:547-55. [Crossref] [PubMed]
- Yeh BW, Li WM, Li CC, et al. Histone deacetylase inhibitor trichostatin A resensitizes gemcitabine resistant urothelial carcinoma cells via suppression of TG-interacting factor. Toxicol Appl Pharmacol 2016;290:98-106. [Crossref] [PubMed]
- Cao QF, Qian SB, Wang N, et al. TRPM2 mediates histone deacetylase inhibition-induced apoptosis in bladder cancer cells. Cancer Biother Radiopharm 2015;30:87-93. [Crossref] [PubMed]
- Li QQ, Hao JJ, Zhang Z, et al. Histone deacetylase inhibitor-induced cell death in bladder cancer is associated with chromatin modification and modifying protein expression: A proteomic approach. Int J Oncol 2016;48:2591-607. [PubMed]
- Schneider AC, Heukamp LC, Rogenhofer S, et al. Global histone H4K20 trimethylation predicts cancer-specific survival in patients with muscle-invasive bladder cancer. BJU Int 2011;108:E290-6. [Crossref] [PubMed]
- Cui J, Sun W, Hao X, et al. EHMT2 inhibitor BIX-01294 induces apoptosis through PMAIP1-USP9X-MCL1 axis in human bladder cancer cells. Cancer Cell Int 2015;15:4. [Crossref] [PubMed]
- Song Y, Wu F, Wu J. Targeting histone methylation for cancer therapy: enzymes, inhibitors, biological activity and perspectives. J Hematol Oncol 2016;9:49. [Crossref] [PubMed]
- You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer cell 2012;22:9-20. [Crossref] [PubMed]
- Shang D, Liu Y, Matsui Y, et al. Demethylating Agent 5-Aza-2′-Deoxycytidine Enhances Susceptibility of Bladder Transitional Cell Carcinoma to Cisplatin. Urology 2008;71:1220-5. [Crossref] [PubMed]
- Karam JA, Fan J, Stanfield J, et al. The use of histone deacetylase inhibitor FK228 and DNA hypomethylation agent 5-azacytidine in human bladder cancer therapy. Int J Cancer 2007;120:1795-802. [Crossref] [PubMed]
- Cheung EM, Quinn DI, Tsaowei DD, et al. Phase II study of vorinostat (Suberoylanilide Hydroxamic Acid, SAHA) in patients with advanced transitional cell urothelial cancer (TCC) after platinum-based therapy—California Cancer Consortium/University of Pittsburgh NCI/CTEP-sponsored trial. J Clin Oncol 2008;26:431-6. [Crossref]
- Wang D, Jing Y, Ouyang S, et al. Inhibitory effect of valproic acid on bladder cancer in combination with chemotherapeutic agents in vitro and in vivo. Oncol Lett 2013;6:1492-8. [PubMed]
- Longo T, McGinley KF, Freedman JA, et al. Targeted Exome Sequencing of the Cancer Genome in Patients with Very High-risk Bladder Cancer. Eur Urol 2016;70:714-7. [Crossref] [PubMed]


