
Extracellular vesicles and their diagnostic and prognostic potential in cancer
Introduction
Cells release a large number of extracellular vesicles (EVs) that carry cargoes, including membrane proteins, cytosolic proteins, mRNAs, and non-coding RNAs such as miRNAs. EVs play a key role in intercellular communication (1-3). Accumulating evidence suggests that tumor-derived EVs play an important role in communication between tumors and their microenvironment (4,5).
Tumor-derived EVs may be a great source of non-invasive cancer biomarkers. EVs can be isolated from many body fluids using non-invasive or minimally invasive procedures. The composition of EVs may reflect that of parental cells and the status of cancer progression. Moreover, EVs are very stable under various storage conditions for a long time, and their lipid bilayers protect macromolecules on and inside EVs from degradation under non-physiological conditions. All of these characteristics qualify tumor-derived EVs as potential diagnostic and prognostic cancer biomarkers (6,7).
EVs are generally classified into two groups based on their sub-cellular origin: exosomes and microvesicles. Exosomes are generated inside the cell during the formation of multivesicular bodies (MVBs). MVBs are late endosomes loaded with intraluminal vesicles (ILVs); ILVs are formed by budding of the limiting membrane of the late endosomes that become MVBs. When exocytosis of MVBs occurs, these ILVs are released to the extracellular space and become exosomes. Microvesicles, on the other hand, are formed and released by outward budding from the plasma membrane. Microvesicles are also called ectosomes, microparticles, and shedding vesicles, among other names (2,8).
Due to the heterogeneous nature of vesicles present in the extracellular environment, the overlapping physical properties of EVs, and the difficulty in determining their sub-cellular origins, classification of EVs has been challenging in practice (2,3,9). Kowal et al. conducted an extensive quantitative proteomic analysis comparing EVs isolated by differential centrifugation and showed that several classical exosome markers are similarly present in all EVs. Moreover, the widely referred exosome fraction prepared by sedimentation at 100,000 g contained EVs that were not associated with the endosomal pathways (10). In fact, biomarkers assigned to exosomes may be either truly exosome-specific or may apply generically to all EVs. In this review, we thus use the generic term “EVs” regardless of the terms used in the articles, but provide detailed information about how authors isolated or captured EVs.
Isolation of EVs
Due to the lack of standardization in the isolation and detection techniques used, many results in the literature are difficult to compare directly. The International Society of Extracellular Vesicles (ISEV) published several position papers that aimed to standardize the protocols used in isolating EVs and their downstream applications (9,11,12). To date, there is no single reliable method for purifying and differentiating exosomes or microvesicles. It is important to take into account the different methods applied when interpreting results. At the end of this section, we summarize a few articles that directly compare multiple EV isolation methods.
Differential centrifugation
Differential centrifugation is the most commonly used method for isolating smaller EVs. The method consists of three steps: (I) depleting cells, dead cells, and cell debris using low-speed centrifugation; (II) removing larger vesicles using centrifugation between 10,000 and 20,000 ×g; and (III) precipitating smaller EVs (often designated as exosome fractions) using ultracentrifugation at 100,000 ×g or higher (13).
The exosomal pellet isolated using differential centrifugation contains various contaminants, such as protein aggregates, low-density lipoprotein (LDL) particles, etc. To improve the purity, an additional density gradient step using sucrose or iodixanol gradients is often applied. Density gradient centrifugation can separate low-density EVs from high-density protein aggregates (13).
Because different types of EVs have similar sedimentation properties, the differential centrifugation method often leads to low purity and unsatisfactory yield. In addition, this method is sensitive to sample viscosity, rotor type, tube k-factor, and centrifugation time (14,15). Unfortunately, identical protocols are often applied to different samples, leading to differences in data obtained by different research groups. Moreover, although straightforward and frequently used, the whole process can be very time-consuming, especially when combined with density gradient procedures. Furthermore, the method requires specialized equipment—an ultracentrifuge—which might not be available in clinical laboratory settings.
Polymer-based precipitation
Another popular protocol uses volume-excluding polymers [e.g., polyethylene glycol (PEG), dextrans, or polyvinyls] or various commercially available polymer-based reagents (e.g., ExoQuick from SBI, Total Exosome Isolation Reagent from Thermo Fisher, and miRCURY from Exiqon, among others). The method generally consists of combining body fluids with a precipitation solution, incubating the resulting mixture overnight at 4 °C, and centrifuging the mixture at low speed to obtain the EV pellet.
A recent method development study of ExtraPEG is worth mentioning (16). Rider et al. tested increasing concentrations of PEG (between 5% and 12%) for their ability to isolate EVs from culture media. They found that ExtraPEG (8% PEG + wash) yielded EVs highly comparable to those obtained using the differential centrifugation method and superior to ExoQuick and Total Exosome Isolation reagents.
These polymer-based reagents are easy to use, need less hands-on time, and do not require special equipment such as an ultracentrifuge. However, these methods may precipitate non-vesicular contaminants, including protein aggregates and lipoproteins, which contain proteins and multiple RNA populations. Even with this limitation, due to their ease of use, polymer-based precipitation methods are becoming increasingly popular.
Immunoaffinity isolation
It is believed that specific surface proteins are present on subsets of EVs, which allows a more restricted population of EVs to be isolated using immunoaffinity isolation methods. In this approach, antibodies against specific surface proteins are covalently attached to beads or other matrices. The most commonly targeted surface proteins are tetraspanins including CD9, CD63, and CD81, and tumor-associated proteins, such as epithelial cell adhesion molecules (EpCAM). This approach may isolate EVs with higher purity and specificity, but may lead to low yield due to its selectivity. In addition, the approach relies heavily on prior knowledge of EV protein content and on highly specific antibodies that may not be readily available.
Importantly, Kowal et al. showed that the 100,000 g pellet from differential centrifugation can be further divided into several categories based on their enrichment or devoid of some or all three tetraspanins, CD63, CD9, and CD81. Therefore, when combined with differential centrifugation, immunoaffinity isolation may be used to enrich specific subtypes of EVs (10).
Size exclusion chromatography (SEC)
SEC allows different sizes of EVs to be eluted from a single column. SEC is relatively quick and can be used for low-volume samples. Importantly, SEC effectively separates EVs from the bulk of soluble proteins. EVs isolated by SEC are minimally altered compared to when precipitating agents or the ultracentrifugation method are used. However, one drawback of this approach is that the final product is often in a large elution volume, and therefore may need to be further concentrated. Moreover, as the separation is purely based on size, some particles such as lipoproteins may be co-isolated. The Exosome Isolation Kit from Evomic Science and qEV from Izon both isolate EVs using SEC principles, but they are not widely cited in the literature.
Filtration
Another method to isolate EVs based on their size is to use ultrafiltration membranes. Filtration is often used in combination with differential centrifugation to remove large particles or vesicles. For example, several studies used 220 nm filtration before differential centrifugation to isolate smaller EVs (17-19).
Other methods
The recently developed polyethylene glycol/dextran aqueous two-phase system has shown to provide high EV recovery efficiency (~70%) in a very short time (15 min). PRotein Organic Solvent PRecipitation (PROSPR) is a new, feasible EV-purification protocol based on protein precipitation by cold acetone (20). Although not used extensively, many researchers have developed different microfluidic devices, such as size-based microfluidics, immunoaffinity-based microfluidic separation, and dynamic microfluidics. The advantages of microfluidics are their sample efficiency, fast separation, and on-chip detection.
Several commercial kits using different EV isolation principles have also been developed. The MagCapture Exosome Isolation Kit PS from Wako Chemicals USA isolates EVs by affinity capturing phosphatidylserine (PS) on the EV using magnetic beads and PS-binding proteins. This kit does not yet have wide application, but it may potentially result in higher yields than most protein-based immunoaffinity isolation methods because it relies on binding lipids rather than proteins. The captured EVs are eluted from beads with metal-chelating reagent at a neutral pH, so the kit potentially enables EVs to be isolated as intact forms. One caveat is that the kit cannot be used for plasma treated with chelating agents, such as EDTA or citric acid and its yield is lower for plasma treated with heparin.
EV isolation kits from Norgen Biotek use patented silicon carbide resin to bind and capture EV membrane proteins. Once captured and precipitated, the supernatant is decanted and EVs are released into the solution with aid of a specialized ‘EV release’ buffer. Ymir Genomics, an early-stage firm, has developed two high-yield methods for isolating EVs from urine. One is based on a novel proprietary molecule that causes EVs to precipitate, while the other is based on a widely available capture resin. The PureExo Exosome Isolation Kit from 101bio.com uses an organic solvent phase partitioning method. Qiagen’s exoEasy kit uses a membrane-based affinity binding step to quickly isolate EVs (21). Its exact affinity binding principle is not clear. The method does not distinguish EVs by size, and does not depend on the presence of a particular surface protein, therefore, it is essential to completely remove cells, dead cells, or apoptotic bodies, etc. in starting materials.
Direct comparison among different EV isolation methods
Different isolation methods are based on different principles and therefore enrich for different subpopulations of EVs. Large-scale studies directly comparing the effects of different isolation methods on EVs and the downstream analyses are lacking. A couple of studies have compared a limited number of techniques and are summarized here.
Helwa et al. compared differential centrifugation and three commercially available polymer-based precipitation reagents (ExoQuick, Total Exosome Isolation Reagent, and miRCURY) for isolation of EVs from serum (22). The three commercial kits consistently had higher yields and higher CD9 and CD63 protein levels than differential centrifugation. However, this difference in EV yields and protein levels did not appear to affect the amount of extracted EV RNA. Importantly, miRNA concentrations varied greatly within and among the different techniques and initial serum volumes used. This miRNA yield variability underscores the importance of using a consistent EV isolation technique and procedure across different samples within the same study. It also illustrates the difficulty of comparing results from different studies using different EV isolation techniques.
Gámez-Valero et al. compared SEC using the Sepharose CL-2B, PEG-based (50% PEG6000), and PROSPR methods for isolation of EVs from plasma (23). Cryo-EM analysis showed the presence of 80–200 nm vesicles in the EVs isolated using SEC; the images also showed very few contaminant particles. In contrast, the authors were not able to visualize any vesicles in the PEG-based preparation, except for some dense contaminant aggregates. PROSPR-prepared EVs appeared to be merged in concentric multi-layer vesicles, which could explain why PROSPR-prepared EVs had many more particles that were larger than 500 nm. Another important observation was that although the protein content of the SEC preparation was extremely low because most of the soluble proteins were eluted in later fractions, PROSPR and PEG preparations had very high protein content, suggesting that these two methods pelleted high quantities of soluble proteins. The authors suggested that the PROSPR and PEG methods may have hampered proteomic analysis of EVs. Lobb et al. drew a similar conclusion that SEC outperforms precipitation methods (24).
Detection and characterization of EVs
Electron microscopy (EM)
Three types of EM have been used to characterize EVs: transmission EM (TEM), scanning EM (SEM), and Cryo-EM. Among them, TEM is most commonly used to obtain size and morphology characteristics of EVs. TEM transmits a beam of electrons through a thin specimen and then focuses the electrons to create an image on a screen. When combined with immunogold labeling, TEM can provide surface protein information of these EVs. As TEM requires fixation and dehydration, it changes the morphology of EVs and may reduce their size.
SEM, although it has lower resolution than TEM, generates detailed three-dimensional images of EV surfaces. Cryo-EM analyzes EVs at temperatures below –100 °C. The main advantage of cryo-EM over TEM and SEM is that samples are vitrified and analyzed without needing to be stained or fixed, which may better preserve EV morphology and characteristics. Although widely used, EM provides limited information on the concentration of the EV preparation.
Atomic force microscopy (AFM)
In AFM, a mechanical probe scans the sample’s surface without actual contact. This ability allows AFM to image soft samples, such as EVs, without damaging them in different environmental media (dried or in liquid), while also providing three-dimensional measurements.
Nanoparticle tracking analysis (NTA)
NTA sizes and counts EVs. It utilizes the properties of light scattering and Brownian motion to obtain the particle size distribution of EVs in liquid suspension. It consists of a laser light scattering microscope, a charge coupled device (CCD) camera that captures the movement of EVs over a series of frames, and proprietary software that tracks many particles individually and calculates their hydrodynamic diameters using the Stokes-Einstein equation. NTA in its fluorescence mode can also be used to analyze fluorescently labeled EVs. As NTA cannot distinguish EVs from other particles such as protein aggregates that may have similar sizes, it is important to isolate EVs prior to NTA analysis.
Tunable resistive pulse sensing (TRPS)
One relatively new technology, qNano from Izon, employs TRPS. qNano detects EVs passing through a nanopore between two chambers by a way of single-molecule electrophoresis. As particles pass through the pore, a transient change in ionic current flow is detected and used to calculate the volume of each particle.
Dynamic light scattering (DLS)
DLS determines the relative size distribution in a fluid relative to the standard population. DLS analysis gives the average size of relatively monodisperse populations of EVs. Because most EV preparation methods result in a heterogeneous mixture of EVs, DLS is less suited in most cases (12).
Zeta potential measurement
EVs bear surface charges and move in an electrical field. The Zeta potential measures this electrophoretic mobility of EVs. The higher the Zeta potential, the less likely that the EVs have aggregated. NTA and RPS technologies have been combined with measurement of the Zeta potential. For example, ZetaView from Particle Metrix can select three measurement modes: size, Zeta potential, and concentration.
Flow cytometry (FC)
FC is very popular method for characterizing EVs, as it can quantify and phenotype large number of EVs in a high-throughput manner. Conventional flow cytometers utilize laser scattering to measure particles. Due to its lower detection limit for light scattering (~300–500 nm), conventional FC has not been suitable for analyzing small EVs. However, over the years, advances in protocols and instruments have allowed researchers to overcome this challenge.
Bead-based methods have been developed for analyzing EVs by FC (25,26). This method consists of three steps: (I) attaching EVs to beads; (II) fluorescently labeling bead-bound EVs, either using fluorochrome-conjugated primary antibodies or fluorochrome-conjugated secondary antibodies (in the latter case, bead-bound EVs are first incubated with primary antibodies); and (III) analyzing with a conventional flow cytometer. The advantages of this approach are that it does not need an advanced or dedicated flow cytometer, and it can determine the relative presence of various surface proteins. However, EV detection largely depends on the abundance or availability of antigens on the EVs and the specificity of the antibodies used. In addition, this method does not quantify EVs.
The research group of Prof. Marca Wauben developed a reliable, high-resolution FC method that can analyze bead-free EVs (27). This protocol uses ultracentrifugation coupled with sucrose gradient flotation for EV isolation, labeling with the lipophilic dye PKH67, and/or fluorochrome-conjugated antibodies. The method has been optimized for the BD Influx flow cytometer (Becton Dickinson) and allows quantification of EVs and further identification of different vesicle subsets expressing particular proteins. This protocol demands an experienced FC operator capable of manual hardware adjustments and calibration to ensure optimized settings are used across experiments. Moreover, the EV labeling step of the experimental procedure, which requires a lengthy sucrose gradient step, is rather time- and labor-intensive.
Pospichalova et al. presented a simplified protocol for direct FC analysis of bead-free EVs using a commercially available dedicated flow cytometer, the Apogee A50 Micro (Apogee Flow Systems) Hertfo (28). EVs can be fluorescently labeled with protein- and/or lipid-specific dyes, without needing to remove the unbound fluorescent dyes in the lengthy sucrose gradient step.
Lannigan and Erdbruegger showed that Imaging Flow Cytometry (ImageStream X MKII, EMD Millipore) can address many limitations encountered using conventional flow cytometry (29). The ImageStream X uses CCD cameras for signal detection, and has a large dynamic range and significantly lower noise. The ImageStream X requires samples to be concentrated into small volumes to collect numbers of events similar to those collected by conventional flow cytometry.
Vesicle flow cytometry (VFC) has recently been developed by Stoner et al. (30). The method involves labeling EVs by a fluorescent lipid probe (di-8-ANEPPS), followed by single particle analysis using a custom-made high sensitivity flow cytometer. Vesicle diameter is estimated by comparison to di-8-stained liposomes (120 nm mean diameter). Since the fluorescent signal is enhanced when the lipid probe is bound and inserted into the outer leaflet of a bilayer membrane, the fluorescent signal is highly vesicle-specific. This approach has the potential to accurately count EVs, as it avoids counting non-vesicle contaminants such as lipoproteins and protein aggregates. However, because this method relies on liposomes (which are heterogeneous in size), it may be less accurate in determining EV size compared to NTA or TRPS. In addition, it cannot detect surface proteins.
Western blotting
This conventional method is widely used to determine the presence or absence of the protein of interest and to detect the level of expression of a selected protein in an EV preparation. Commonly used protein markers are CD9, CD63, CD81, TSG101, Alix, actin, tubulin, flotillin-1, HSC70/HSP73, HSP70/HSP72, and MHC molecules. Western blotting of these markers, however, cannot be used to quantify EVs and the enrichment of these proteins in the EV fraction does not guarantee the absence of contaminants. The absence of cell-derived organelle markers such as calreticulin is often used to assess the purity of an EV preparation (12).
Other methods
Raman spectroscopy has also been used to characterize EVs. However, conventional Raman spectroscopy is a very inefficient process and requires high sample concentration, high laser power, and long signal integration times. Stremersch et al. presented a proof-of-concept in which they used Surface Enhanced Raman Spectroscopy (SERS) to identify single EVs (31). Each individual EV coated with gold nanoparticles generates an optical fingerprint, which may hold great potential for identifying and classifying different subtypes of EVs.
Comparative analysis
Akers et al. recently compared the abilities of TEM, NTA, TRPS, and VFC to quantify EVs from clinical cerebrospinal fluid (CSF) (32). When TEM was used, the particle count was consistently two orders of magnitude lower than the counts generated by NTA and TRPS. VFC particle counts were consistently two to three times lower than those of NTA and TRPS, possibly due to the fact that NTA and TRPS counted protein aggregates or other non-vesicle particles in addition to EVs. Interestingly, NTA and TRPS differed in counting EVs of different sizes. For EVs <150 nm in diameter, NTA detected more EVs than TRPS, while for EVs >150 nm in diameter, NTA detected fewer EVs than TRPS. Thus, NTA was better at detecting small EVs, while TRPS excelled at detecting EVs that were larger.
Furthermore, van der Pol et al. compared TEM, NTA, TRPS, conventional FC, and dedicated FC (Apogee A50 Micro) for quantifying EVs from urine and polystyrene beads (33). Similar to the report by Akers et al., each technique gave different particle size distributions for the same sample. In terms of concentration measurements, NTA and TRPS were similar. Compared to NTA and TRPS measurements, conventional FC underestimated the concentration almost 300-fold, and the more sensitive, high-resolution flow cytometer underestimated the EV concentration 15-fold. Interestingly, the TEM-measured concentration was 7–8 times less than NTA or TRPS measurements, suggesting that TEM’s measurement of concentration is highly variable due to the non-homogenous attachment of EVs to the EM grid.
Maas et al. compared NTA, TRPS, and high-resolution FC for analysis of cell culture EVs, polystyrene beads, and calcein-loaded liposomes (34). The authors found that variables such as particle concentration range, the NTA camera level and detection threshold, and the nanopore size in TRPS significantly influenced the measurements. In addition, they found bigger differences in terms of EV counting for NTA and TRPS. The differences between this study and those of van der Pol et al. and Akers et al. may be due to the fact that the other two studies did not change the default settings of the commercial instruments. Together, these studies emphasize the importance of standardizing EV characterization procedures, so more rigorous comparative studies like these can be made.
EVs as cancer biomarkers
It has been well established that various tumors can shed intact tumor cells [resulting in circulating tumor cells (CTCs)], tumor-derived EVs, and cellular components such as nucleic acids [resulting in cell-free DNA (cfDNA) or RNA] in body fluids. In fact, CTCs, cfDNAs, and EVs are being studied as potential sources for liquid biopsy. The key advantages of EVs as liquid biopsy material are fourfold. First, compared cfDNAs, EVs are extremely stable and can keep their contents intact for a much longer time than other materials. Second, EVs, like CTCs, allow analyses of proteins in addition to nucleic acid sequences. Third, compared to CTCs, EVs are much more abundant, allowing further manipulation. Fourth, although CTCs have potential as prognostic markers of metastatic development, incurable metastasis can already be developed at the time of initial diagnosis with existing CTC assays. In contrast, tumor-derived EVs may be a more valuable prognostic marker because they are released from the primary tumor long before detectable CTCs are present in body fluids.
EV cargo profiling and characterization for biomarker discovery
Molecular profiling and characterization of EV cargoes is of significant interest because they can provide clues about EV biogenesis and cellular function, and can be used to develop biomarkers for disease diagnosis, prognosis, and treatment response. EVs carry membrane proteins, cytosolic proteins, mRNAs, and non-coding RNAs such as miRNAs. Among these, EV miRNAs are best studied. Overall, there are fewer EV protein cargo papers than EV RNA papers, due to the lack of amplification procedures for identifying protein cargoes.
There are generally two ways to discover EV protein cargoes as potential biomarkers. First, proteomics analyses using mass spectrometry in disease-related cell lines or patient samples have been conducted to find candidate protein cargoes. Next, the question of whether the candidate protein is a “real” EV cargo is answered using immunogold TEM, Western blotting, or flow cytometry. Lastly, EV protein cargoes have been tested in patient samples to determine their diagnostic or prognostic potential using flow cytometry, mass spectrometry, or enzyme-linked immunosorbent assay (ELISA). Another method is to phenotype the known surface proteins on EVs using protein microarrays or ELISA; however, this method depends greatly on the availability of highly-specific antibodies and prior knowledge of EV protein cargo compositions.
EV RNA biomarkers can be discovered in two different ways. In the first procedure, EV RNAs are profiled using RNA microarray or RNA sequencing, their differential expressions in patients and healthy donors are compared, and potential RNA markers are thus identified. Then, the candidates are tested using RT-qPCR to determine their clinical potential as biomarkers. In the second method, known miRNA species are tested for their differential expression using RT-qPCR to determine their clinical potential.
EV proteins as diagnostic and prognostic biomarkers
Table 1 summarizes the EV proteins that have been shown to have diagnostic or prognostic potential in various cancers. Melo et al. showed that proteoglycan glypican-1 (GPC1)-positive EVs were detected in the serum of patients with pancreatic cancer with absolute specificity and sensitivity, distinguishing them from healthy subjects and patients with a benign pancreatic disease (17). Levels of GPC1-positive EVs have correlated with tumor burden and survival of pre- and post-surgical patients, suggesting a prognostic relevance of the marker. A recent publication by Lai et al., however, concluded that EV GPC1 is not diagnostic for pancreatic cancer (18). The authors used a liquid chromatography-tandem mass spectrometry (LC-MS/MS) method to quantitatively analyze EV GPC1. They assessed EV GPC1 levels from normal controls, three patients with chronic pancreatitis (CP), and three patients with pancreatic ductal adenocarcinoma (PDAC) and found no statistical difference in EV GPC1 level. Moreover, EV GPC1 levels only slightly decreased after PDAC resection. Their procedure for isolating EVs was similar to the one reported by Melo et al. One caveat of the Lai et al. study is that the numbers of patients in each category is small. Another possibility for the result, as discussed by Lai et al., is that the antibody used in Melo et al. may recognize specific glycosylated cancer-specific epitopes of GPC1, while the LC-MS/MS method used in the latter study detected only core GPC1 proteins. The potential of GPC1 as a diagnostic and prognostic marker for pancreatic cancer needs further validation.
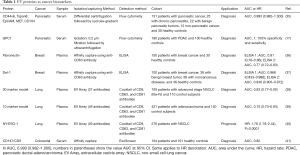
Full table
The above-mentioned studies, although exciting, raise several issues, including the applicability of the methods to the clinical setting, as the authors used lengthy ultracentrifugation procedures to isolate EVs. In fact, several groups aim to skip the EV isolation step and quickly capture and detect EVs to determine the relative abundance of EV surface proteins as potential biomarkers. After using proteomic studies to identify potential biomarkers, Moon et al. developed ELISA methods to capture and determine two proteins that may be used as diagnostic biomarkers in breast cancer. The authors used two different antibodies for each protein and obtained similar AUC and other clinically relevant parameters (36,37). One of the biomarkers, Del-1, showed AUCs of 0.968 and 0.946 in two different ELISA assays, suggesting that EV Del-1 could be a promising marker for breast cancer detection (37).
Three studies used EV arrays to identify potential cancer biomarkers. In these studies, 37 or 49 antibodies were printed on an array and used to capture EVs expressing specific surface proteins, then the captured EVs were detected using a cocktail of three antibodies against CD9, CD63, and CD81. This method successfully identified a 30-marker model and a 10-marker model for diagnosing advanced non-small cell lung cancer (NSCLC) and adenocarcinoma, respectively, and also NY-ESO-1 as a potential prognostic marker for NSCLC (38-40).
Another study used ExoScreen, a novel method that specifically detects EVs in the serum based on an amplified luminescent proximity homogeneous assay using photosensitizer beads that can detect EVs without a prior isolation step. The study found that CD147 and CD9 double-positive EVs were significantly higher in serum from colorectal cancer patients than in serum from healthy donors (AUC 0.82) (41).
EV miRNAs as diagnostic and prognostic biomarkers
miRNAs are short, single-stranded, non-coding RNA molecules comprised of 19–22 nucleotides. The latest miRBase release (June 2013) contains more than 2,500 mature human miRNA sequences (42). Many miRNAs are dysregulated in various cancers, and their differential expression profiles are associated with tumor development, differentiation, progression, and response to therapy. Importantly, miRNAs can be secreted from cells and taken up by other cells. Cell-free circulating miRNAs have been found in various human body fluids including breast milk, colostrum, saliva, seminal fluid, tears, urine, amniotic fluid, CSF, blood, pleural fluid, peritoneal fluid, and bronchial lavage fluid (43).
Table 2A,B summarizes EV miRNAs that have been shown to have diagnostic or prognostic potential in various cancers. The results of these studies are difficult to compare directly due to many factors. First, the sample types from which the EVs were derived are different. In a 2017 position paper, ISEV recommended that plasma rather than serum should be used for EV nucleic acid analysis because serum contains high numbers of EVs released by platelets in response to coagulation, while plasma mainly contains EVs originally present in the circulating blood (9). Second, different EV isolation methods were employed. Many studies used polymer-based precipitation methods, which can precipitate many particles, including protein aggregates and lipoproteins. Third, methodological pitfalls that could affect miRNA measurements must be taken into account, such as the RNA isolation techniques, storage conditions, quality control methods, quantification principles, array platforms, library preparation protocols, sequencing methods, RT-qPCR principles, and methods for evaluating data, especially normalization strategies. For example, the popular RNA extraction reagent Trizol has been shown to induce GC content bias in small RNAs (61), and the popular internal control U6 has been reported to be unsuitable as an endogenous control for the quantification of cell-free miRNAs (62,63).

Full table

Full table
Not all circulating miRNAs are from EVs. Besides being packaged into EVs, circulating miRNAs have been found to be associated with ribonucleoprotein complexes (such as Argonaute proteins) (64,65), lipoproteins (66), and nucleophosmin (67), and also exist as free-circulating miRNAs. Although two studies showed that only a minor portion of cell-free miRNAs are associated with EVs (64,68), other studies have shown that expression levels of particular miRNAs are significantly higher in EVs compared to total serum (44,52,69).
Do EV miRNAs have better diagnostic or prognostic potential than miRNAs recovered from whole body fluid? It depends. Uratani et al. performed a head-to-head comparison between miRNA expression levels of total serum and EV fractions, and demonstrated that although the diagnostic potential of both serum miR-21 and EV miR-21 for adenoma patients is similar, serum miR-21 is a superior diagnostic biomarker for advanced, high-risk adenoma (52). Lai et al. identified a number of EV and plasma miRNAs that have diagnostic potential for PDAC. Interestingly, ROC curves showed that plasma miR-106b levels were more sensitive for differentiating PDAC from normal samples than EV miR-106b levels (AUC 0.98 vs. 0.85). On the other hand, in contrast to plasma miR-122, with an AUC of 0.89, EV miR-122 had an AUC of 0.99 (18). Therefore, cell-free circulating miRNAs and EV miRNAs may have different diagnostic or prognostic potentials in different cancers. More comparative analyses are necessary to identify the best biomarkers for various cancers.
Another interesting study examined EV miR-21 expression in both the serum and CSF of glioma patients (54). The EV miR-21 level in CSF appeared to be an excellent index to differentiate glioma from non-tumor brain diseases (AUC 0.927; 95% CI, 0.865–0.985), and had good potential for discriminating grade III/VI from grade II gliomas (AUC 0.872; 95% CI, 0.817–0.927). Interestingly, EV miR-21 levels in serum were not different between glioma patients and controls. Thus, tumor-related body fluids may have advantages for identifying EV-derived biomarkers.
Particular isoforms of EV miRNAs may be better cancer biomarker candidates in some cases. Koppers-Lalic et al. observed that miRNA isoforms with 3’ end modifications of miR-21, miR-204, and miR-375 were highly discriminatory with respect to urine EV samples from control men vs. prostate cancer patients. On the other hand, the mature forms of these three miRNAs in urine EV samples failed to robustly discriminate for disease (45).
Other EV RNAs as diagnostic and prognostic biomarkers
The clinical utility of EV RNA (not EV miRNA) is also under investigation. Wang et al. showed that combined expression of miR-21 and HOTAIR from serum EV is superior to EV miR-21 or EV HOTAIR alone in differentiating malignant from benign laryngeal squamous cell carcinoma (70). Donovan et al. showed that the gene signature derived from normalized PCA3 and ERG RNA expression in EVs from urine can predict initial biopsy results in prostate cancer patients (59). A subsequent prospective study confirmed the utility of this assay (60). This non-invasive assay is now commercialized by Exosome Diagnostics.
Conclusions
Tumor-derived EVs and EV cargoes are highly promising biomarker candidates for the diagnosis and prognosis of cancers. Current methods for isolating and characterizing EVs, as well as for detecting EV cargoes, are highly variable. Robust comparative analyses would help develop standardized methods that would enable more reliable inter-study validation of EV cargoes as cancer biomarkers.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.06.32). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hopkin K. Core Concept: Extracellular vesicles garner interest from academia and biotech. Proc Natl Acad Sci 2016;113:9126-8. [Crossref] [PubMed]
- Tkach M, Théry C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 2016;164:1226-32. [Crossref] [PubMed]
- Maas SLN, Breakefield XO, Weaver AM. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol 2017;27:172-88. [Crossref] [PubMed]
- Wendler F, Favicchio R, Simon T, et al. Extracellular vesicles swarm the cancer microenvironment: from tumor-stroma communication to drug intervention. Oncogene 2017;36:877-84. [Crossref] [PubMed]
- Becker A, Thakur BK, Weiss JM, et al. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell 2016;30:836-48. [Crossref] [PubMed]
- Kinoshita T, Yip KW, Spence T, et al. MicroRNAs in extracellular vesicles: potential cancer biomarkers. J Hum Genet 2017;62:67-74. [Crossref] [PubMed]
- Xu R, Greening DW, Zhu HJ, et al. Extracellular vesicle isolation and characteriza-tion: toward clinical application. J Clin Invest 2016;126:1152-62. [Crossref] [PubMed]
- Cocucci E, Meldolesi J. Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol 2015;25:364-72. [Crossref] [PubMed]
- Mateescu B, Kowal EJ, van Balkom BW, et al. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA – an ISEV position paper. J Extracell Vesicles 2017;6:1286095 [Crossref] [PubMed]
- Kowal J, Arras G, Colombo M, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A 2016;113:E968-77. [Crossref] [PubMed]
- Hill AF, Pegtel DM, Lambertz U, et al. ISEV position paper: extracellular vesicle RNA analysis and bioinformatics. J Extracell Vesicles 2013;2. [PubMed]
- Witwer KW, Buzás EI, Bemis LT, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles 2013;2. [PubMed]
- Théry C, Amigorena S, Raposo G, et al. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol 2006;Chapter 3:Unit 3.22.
- Cvjetkovic A, Lötvall J, Lässer C. The influence of rotor type and centrifugation time on the yield and purity of extracellular vesicles. J Extracell Vesicles 2014;3. [PubMed]
- Livshits MA, Khomyakova E, Evtushenko EG, et al. Isolation of exosomes by differential centrifugation: Theoretical analysis of a commonly used protocol. Sci Rep 2015;5:17319. [Crossref] [PubMed]
- Rider MA, Hurwitz SN, Meckes DG. ExtraPEG: A Polyethylene Glycol-Based Method for Enrichment of Extracellular Vesicles. Sci Rep 2016;6:23978. [Crossref] [PubMed]
- Melo SA, Luecke LB, Kahlert C, et al. Glypican1 identifies cancer exosomes and facilitates early detection of cancer. Nature 2015;523:177-82. [Crossref] [PubMed]
- Lai X, Wang M, McElyea SD, et al. A microRNA signature in circulating exosomes is superior to exosomal glypican-1 levels for diagnosing pancreatic cancer. Cancer Lett 2017;393:86-93. [Crossref] [PubMed]
- Que R, Ding G, Chen J, et al. Analysis of serum exosomal microRNAs and clinicopathologic features of patients with pancreatic adenocarcinoma. World J Surg Oncol 2013;11:219. [Crossref] [PubMed]
- Gallart-Palau X, Serra A, Wong AS, et al. Extracellular vesicles are rapidly purified from human plasma by PRotein Organic Solvent PRecipitation (PROSPR). Sci Rep 2015;5:14664. [Crossref] [PubMed]
- Enderle D, Spiel A, Coticchia CM, et al. Characterization of RNA from Exosomes and Other Extracellular Vesicles Isolated by a Novel Spin Column-Based Method. PLoS One 2015;10:e0136133 [Crossref] [PubMed]
- Helwa I, Cai J, Drewry MD, et al. A Comparative Study of Serum Exosome Isolation Using Differential Ultracentrifugation and Three Commercial Reagents. PLoS One 2017;12:e0170628 [Crossref] [PubMed]
- Gámez-Valero A, Monguió-Tortajada M, Carreras-Planella L, et al. Size-Exclusion Chromatography-based isolation minimally alters Extracellular Vesicles’ characteristics compared to precipitating agents. Sci Rep 2016;6:33641. [Crossref] [PubMed]
- Lobb RJ, Becker M, Wen SW, et al. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J Extracell Vesicles 2015;4:27031. [Crossref] [PubMed]
- Dragovic RA, Collett GP, Hole P, et al. Isolation of syncytiotrophoblast microvesicles and exosomes and their characterisation by multicolour flow cytometry and fluorescence Nanoparticle Tracking Analysis. Methods 2015;87:64-74. [PubMed]
- Inglis H, Norris P, Danesh A. Techniques for the Analysis of Extracellular Vesicles Using Flow Cytometry. J Vis Exp 2015;52484. [PubMed]
- van der Vlist EJ, Nolte-’t Hoen EN, Stoorvogel W, et al. Fluorescent labeling of nanosized vesicles released by cells and subsequent quantitative and qualitative analysis by high-resolution flow cytometry. Nat Protoc 2012;7:1311-26. [Crossref] [PubMed]
- Pospichalova V, Svoboda J, Dave Z, et al. Simplified protocol for flow cytometry analysis of fluorescently labeled exosomes and microvesicles using dedicated flow cytometer. J Extracell Vesicles 2015;4:25530. [Crossref] [PubMed]
- Lannigan J, Erdbruegger U. Imaging flow cytometry for the characterization of ex-tracellular vesicles. Methods 2017;112:55-67. [Crossref] [PubMed]
- Stoner SA, Duggan E, Condello D, et al. High sensitivity flow cytometry of mem-brane vesicles. Cytometry A 2016;89:196-206. [Crossref] [PubMed]
- Stremersch S, Marro M, Pinchasik BE, et al. Identification of Individual Exosome-Like Vesicles by Surface Enhanced Raman Spectroscopy. Small 2016;12:3292-301. [Crossref] [PubMed]
- Akers JC, Ramakrishnan V, Nolan JP, et al. Comparative Analysis of Technologies for Quantifying Extracellular Vesicles (EVs) in Clinical Cerebrospinal Fluids (CSF). PLoS One 2016;11:e0149866 [Crossref] [PubMed]
- van der Pol E, Coumans FA, Grootemaat AE, et al. Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flow cytometry, nanoparticle tracking analysis, and resistive pulse sensing. J Thromb Haemost 2014;12:1182-92. [Crossref] [PubMed]
- Maas SL, de Vrij J, van der Vlist EJ, et al. Possibilities and limitations of current technologies for quantification of biological extracellular vesicles and synthetic mimics. J Control Release 2015;200:87-96. [Crossref] [PubMed]
- Madhavan B, Yue S, Galli U, et al. Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int J Cancer 2015;136:2616-27. [Crossref] [PubMed]
- Moon PG, Lee JE, Cho YE, et al. Fibronectin on circulating extracellular vesicles as a liquid biopsy to detect breast cancer. Oncotarget 2016;7:40189-99. [PubMed]
- Moon PG, Lee JE, Cho YE, et al. Identification of Developmental Endothelial Locus1 on Circulating Extracellular Vesicles as a Novel Biomarker for Early Breast Cancer Detection. Clin Cancer Res 2016;22:1757-66. [Crossref] [PubMed]
- Jakobsen KR, Paulsen BS, Bæk R, et al. Exosomal proteins as potential diagnostic markers in advanced non-small cell lung carcinoma. J Extracell Vesicles J Extracell Vesicles 2015;4:26659. [Crossref] [PubMed]
- Sandfeld-Paulsen B, Jakobsen KR, Bæk R, et al. Exosomal Proteins as Diagnostic Biomarkers in Lung Cancer. J Thorac Oncol 2016;11:1701-10. [Crossref] [PubMed]
- Sandfeld-Paulsen B, Aggerholm-Pedersen N, Bæk R, et al. Exosomal proteins as prognostic biomarkers in non-small cell lung cancer. Mol Oncol 2016;10:1595-602. [Crossref] [PubMed]
- Yoshioka Y, Kosaka N, Konishi Y, et al. Ultra-sensitive liquid biopsy of circulating extracellular vesicles using ExoScreen. Nat Commun 2014;5:3591. [Crossref] [PubMed]
- Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 2014;42:D68-73. [Crossref] [PubMed]
- Weber JA, Baxter DH, Zhang S, et al. The MicroRNA Spectrum in 12 Body Fluids. Clin Chem 2010;56:1733-41. [Crossref] [PubMed]
- Li Z, Ma YY, Wang J, et al. Exosomal microRNA-141 is upregulated in the serum of prostate cancer patients. Onco Targets Ther 2015;9:139-48. [PubMed]
- Koppers-Lalic D, Hackenberg M, de Menezes R, et al. Non-invasive prostate cancer detection by measuring miRNA variants (isomiRs) in urine extracellular vesicles. Oncotarget 2016;7:22566-78. [PubMed]
- Huang X, Yuan T, Liang M, et al. Exosomal miR-1290 and miR-375 as Prognostic Markers in Castration-resistant Prostate Cancer. Eur Urol 2015;67:33-41. [Crossref] [PubMed]
- Machida T, Tomofuji T, Maruyama T, et al. miR-1246 and miR-4644 in salivary exosome as potential biomarkers for pancreatobiliary tract cancer. Oncol Rep 2016;36:2375-81. [PubMed]
- Zhang W, Ni M, Su Y, et al. MicroRNAs in Serum Exosomes as Potential Bi-omarkers in Clear-cell Renal Cell Carcinoma. Eur Urol Focus 2016. Available online: http://dx.doi.org/
10.1016/j.euf.2016.09.007 - Butz H, Nofech-Mozes R, Ding Q, et al. Exosomal MicroRNAs Are Diagnostic Biomarkers and Can Mediate Cell–Cell Communication in Renal Cell Carcinoma. Eur Urol Focus 2016;2:210-8. [Crossref]
- Matsumura T, Sugimachi K, Iinuma H, et al. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br J Cancer 2015;113:275. [Crossref] [PubMed]
- Liu C, Eng C, Shen J, et al. Serum exosomal miR-4772-3p is a predictor of tumor recurrence in stage II and III colon cancer. Oncotarget 2016;7:76250-60. [PubMed]
- Uratani R, Toiyama Y, Kitajima T, et al. Diagnostic Potential of Cell-Free and Exosomal MicroRNAs in the Identification of Patients with High-Risk Colorectal Adenomas. PLoS One 2016;11:e0160722 [Crossref] [PubMed]
- Ogata-Kawata H, Izumiya M, Kurioka D, et al. Circulating Exosomal microRNAs as Biomarkers of Colon Cancer. PLoS One 2014;9:e92921 [Crossref] [PubMed]
- Shi R, Wang PY, Li XY, et al. Exosomal levels of miRNA-21 from cerebrospinal fluids associated with poor prognosis and tumor recurrence of glioma patients. Oncotarget 2015;6:26971-81. [Crossref] [PubMed]
- Manterola L, Guruceaga E, Pérez-Larraya JG, et al. A small noncoding RNA signature found in exosomes of GBM patient serum as a diagnostic tool. Neuro-Oncol 2014;16:520. [Crossref] [PubMed]
- Jiao C, Jiao X, Zhu A, et al. Exosomal miR-34s panel as potential novel diagnostic and prognostic biomarker in patients with hepatoblastoma. J Pediatr Surg 2017;52:618-24. [Crossref] [PubMed]
- Cazzoli R, Buttitta F, Nicola MD, et al. MicroRNAs derived from circulating exosomes as non-invasive biomarkers for screening and diagnose lung cancer. J Thorac Oncol 2013;8:1156. [Crossref] [PubMed]
- Wang J, Zhou Y, Lu J, et al. Combined detection of serum exosomal miR-21 and HOTAIR as diagnostic and prognostic biomarkers for laryngeal squamous cell carcinoma. Med Oncol 2014;31:148. [Crossref] [PubMed]
- Donovan MJ, Noerholm M, Bentink S, et al. A molecular signature of PCA3 and ERG exosomal RNA from non-DRE urine is predictive of initial prostate biopsy result. Prostate Cancer Prostatic Dis 2015;18:370-5. [Crossref] [PubMed]
- McKiernan J, Donovan MJ, O’Neill V, et al. A Novel Urine Exosome Gene Expression Assay to Predict High-grade Prostate Cancer at Initial Biopsy. JAMA Oncol 2016;2:882-9. [Crossref] [PubMed]
- Kim YK, Yeo J, Kim B, et al. Short Structured RNAs with Low GC Content Are Selectively Lost during Extraction from a Small Number of Cells. Mol Cell 2012;46:893-5. [Crossref] [PubMed]
- Benz F, Roderburg C, Vargas Cardenas D, et al. U6 is unsuitable for normalization of serum miRNA levels in patients with sepsis or liver fibrosis. Exp Mol Med 2013;45:e42 [Crossref] [PubMed]
- Xiang M, Zeng Y, Yang R, et al. U6 is not a suitable endogenous control for the quantification of circulating microRNAs. Biochem Biophys Res Commun 2014;454:210-4. [Crossref] [PubMed]
- Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A 2011;108:5003-8. [Crossref] [PubMed]
- Turchinovich A, Weiz L, Langheinz A, et al. Characterization of extracellular circulating microRNA. Nucleic Acids Res 2011;39:7223-33. [Crossref] [PubMed]
- Vickers KC, Palmisano BT, Shoucri BM, et al. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 2011;13:423-33. [Crossref] [PubMed]
- Wang K, Zhang S, Weber J, et al. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res 2010;38:7248-59. [Crossref] [PubMed]
- Chevillet JR, Kang Q, Ruf IK, et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci U S A 2014;111:14888-93. [Crossref] [PubMed]
- Wang H, Hou L, Li A, et al. Expression of serum exosomal microRNA-21 in human hepatocellular carcinoma. BioMed Res Int 2014;2014:864894 [PubMed]
- Wang J, Zhou Y, Lu J, et al. Combined detection of serum exosomal miR-21 and HOTAIR as diagnostic and prognostic biomarkers for laryngeal squamous cell carcinoma. Med Oncol 2014;31:148. [Crossref] [PubMed]


