Radiogenomics: the utility in patient selection
Introduction
State of art in patient selection for radiotherapy treatment
In recent years the technological development of radiation therapy (RT) has led to more efficient processing techniques which can deliver with high precision, increasing dose savings to the organ at risk (OAR), and high dose values on tumor targets, even of small size. The main innovation in RT starts with the 3D conformal radiation therapy (3D-CRT) (1). This is a technique where the beams of radiation are shaped to target the tumor. The target location is obtained by the CT, MRI and CT/PET exams, used individually or together; then, a software analyzes the 3D image and designs radiation beams that follow the shape and geometry of the tumor. The multi leaf collimator (MLC) permits to deliver radiation beams with irregular and asymmetric geometries in order to irradiate the tumor shape.
The intensity modulated radiation therapy (IMRT) is an advanced type of conformal RT and it uses non-uniform radiation beams with different intensities (2). The modulated beam allows to obtain a painting of the radiation dose to the target volume, in particular with concave or complex shapes. Moreover, it is possible to observe a better dose uniformity at the target and a largest dose saving surrounding healthy tissues.
Another IMRT evolution is the Tomotherapy, a system equipped with a linear accelerator, software and technological elements to perform a modulation of the dose intensity (3). At the same time, there is a system capable of processing CT images, in order to scan the patient treatment position during each session.
Further technological advances in RT are evolving with the use of hadrons, such as protons, neutrons and carbon ions (4). The hadrontherapy is indicated for tumors in which conventional RT does not give significant advantages: in particular, for radio-resistant tumors and for those located close to organs at risk. The use of hadrons allows to irradiate deep tissues and to spare surrounding healthy tissues.
Similarly to IMRT, it has also been developed the intensity modulated proton therapy (IMPT). The IMPT uses “pencil beams” of protons of variable energy and intensity, to create target-local modulations increasing the dose coverage on the target and reducing the dose at the OAR (5).
The goal of RT is to deliver the therapeutic dose to target tissues minimizing the risks of normal tissue complication (6). The latter reason conducted to the development, in the last year, of new technologies in the RT fields in order to improve the capability to deposit the dose in the target volume sparing, as good as possible, the healthy tissue. Usually, during a treatment planning it is not possible to avoid completely the irradiation of healthy tissues and, as a consequence, the treatment planner needs information to predict the risk of a normal tissue injury for optimizing the 3D dose distribution. The first paper published about the healthy tissue tolerance was written by Emami et al. (7). In 2010, Bentzen et al. (8) wrote a paper about the tolerance dose for irradiation of one third, two thirds or the whole of various organs. Due to the scarceness of clinical data, the team members chose the approach to establish the tolerance dose only by using clinical experience.
In order to facilitate the use of the Emami et al. constraints, Kutcher et al. (9) suggested a reduction algorithm called “Dose-Volume Histogram” (DVH) that permits the extrapolation of Emami’s constraint to any dose distribution. With the aim to revise the Emami’s guidelines, the Quantitative Analysis of Normal Tissue Effects in the Clinic (QUANTEC) Steering Committee was formed. This committee defined three aims for the QUANTEC guidelines:
- To provide a critical overview of the current state of knowledge on quantitative dose-response and dose-volume relationships for clinically relevant normal-tissue end-points;
- To produce practical guidance allowing the clinician to reasonably categorize toxicity risk based on dose-volume parameters or model results;
- To identify future research avenues that would help to improve risk estimation or mitigation of early and late side effects of RT.
It was a hard work also because the literature on risk factor related with patients is dispersed and quite often inconsistent from one study to the next. Different validation was performed on the QUANTEC Steering Committee results (8) and today the dose-volume constraints are routinely used during a treatment planning and are considered one of the final tests to perform in order to verify the goodness of a treatment plan. A dose distribution is considered acceptable, not only using numerical estimates from dose-volume models, but also, performing an assessment of the risk-benefit ratio for each single patient, performed based on clinical experience.
Tumor heterogeneity and toxicity effects in RT response
Technological advances in radiation delivery and the introduction of particle therapies have strongly limited the amount of dose distributed to normal tissues and enhanced the tumor killing capacity. Nevertheless, RT treatment planning should take in account biomarkers of normal tissues and tumor radiosensitivity, which are currently studied only at a research level, with the exception of few mutations which are clinically noted to affect the insurgence of radiation toxicity and side effects.
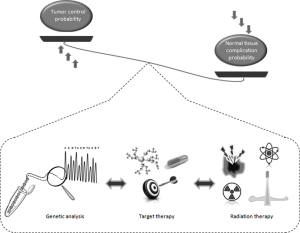
In this post-genomic era, the possibility of including such biomarkers in the treatment planning could be translated in the possibility of optimizing therapeutic efficacy, controlling the normal tissue complication probability (NCTP) and enhancing the tumor control probability (TCP), in a personalised manner. This knowledge could permit to prescribe an increased total dose to the tumor in patients with relatively radioresistant normal tissues and to address candidate patients having high risk of developing severe normal-tissue reaction to either different RT regimens (alternative fractionation schedules, treatment planning, or modalities) or alternative treatments (surgery, chemotherapy, target therapy, ablative treatments, etc.). Today, RT planning is based on physical metrics such as radiation dose and tumor volume, because of these parameters are measurable, and thus the treatment plan is directly verifiable. Otherwise, treatment plan could be choose evaluating biological parameters, including mutations, omic biomarkers, % of cancer stem cells (CSCs), % of hypoxia (eventually extracted by molecular imaging methods), and used in patient stratification and radiation dose prescription.
For example, a recent biological driven approach is based on the spatial dose optimization within the tumor target volume using the dose-painting evaluation. In this way, the more proliferative and metabolically active tumor areas, potentially resistant to RT, could be identified and thus irradiated using an higher dose compared to the other parts of the tumor. In this sense during the course of RT, in several clinical studies the metabolically active tumor areas are selected by fluorodeoxyglucose (FDG) PET imaging as well as the hypoxic tumor subvolumes are selected by FMISO PET or (18F) fluoroazomycin arabinoside (FAZA) PET (10).
Another possibility to produce a biological driven RT treatment planning is to extrapolate measurable parameters, such as the biological response (for example, in term of tumor cell death) rather than to use radiation dose. The response to the first few treatment fractions, assessed with molecular imaging, could be used as parameters to adapt the treatment if the response is above or below the expected value.
These types of approaches could, perhaps, help to drive towards a biological driven treatment plans, overcoming the difficulties to identify radiosensitivity biomarkers unique for all tumor types and organs at risk. The next paragraphs describe the complexity of biological factors conditioning the success of RT and the occurrence of normal tissue complications.
Genomic tumor heterogeneity
Cancers are highly heterogeneous diseases, at both clinical and molecular levels, presenting distinct subtypes associated with different clinical outcomes (11,12). Understanding this heterogeneity represents a key factor for the development of targeted preventive and therapeutic interventions.
The individual response to RT is well established. It depends by intrinsic tumor cell type features, such as germline and acquired mutations, genomic instability, tumor progression mediated by epithelial-mesenchymal transition (EMT), presence of CSCs, etc. (13-15). Extrinsic factors, including dose, age, additional treatment and comorbidities can also modify individual RT response. However, excluding these extrinsic factors, about the 80% of individual variations to RT remains unexplained, raising the possibility of underlying genetic differences as a cause for these variations (16).
A general concept is that DNA damage levels that exceed the repair capacity of tumor and normal cells lead to the activation of apoptosis, which is controlled by a number of proapoptotic and antiapoptotic factors (17). Then, the same pathways are involved both in conditioning the tumor eradication and RT success, thus in TCP, both in the occurrence of normal tissue sensitivity, thus in NTCP. Mutation located on crucial genes belonging to these pathways, globally affect radiation tissue response.
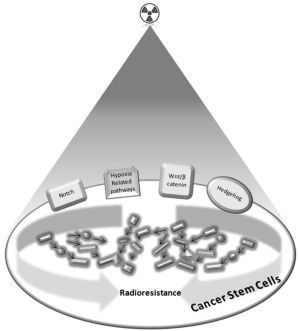
Radiogenomics is the whole genome application which can help to identify these genetic biomarkers regulating radiation sensitivity, in order to reach a maximum TCP with minimum NTCP (Figure 1).
A comprehensive review by Guo and collaborators (17) well synthesize crucial pathways driving the response to ionizing radiations (IR). In particular, the following four classical signal transduction pathways, PI3K/AKT, MAPK/ERK, NF-κB, TGF-β, are key regulators of tumor radiation response. Moreover, these pathways are able to affect the expression of crucial genes in nucleus, which take part in the processes of DNA damage repair, cell cycle progression and apoptosis. Interestingly, candidate genes associated to control radiosensitivity are involved in DNA repair process (ATM, ATR, MDC1, 53BP1, MRE11-RAD50-NBS1 complex, DNA-PKcs, Ku70/Ku80, LIG4, XRCC1, XRCC2, XRCC3, XRCC4, XRCC5, BRCA1, BRCA2, RAD51 and RAD52, NBN); cell cycle control (CDC25, P53, P21, 53BP1); oxidative stress response (TXNRD2, GSTP1 and endothelial nitric oxide synthase) and in apoptosis regulation (FAS, FASL, TP53, TP73 and HDM2) (17).
Thanks to the omics technologies, now is possible to detect disease- or resistance-driving mutations in just one assay, permitting clinicians to define personalized treatments in radiation oncology also in combination with other interventions
Recently, a very interesting review has reported a number of clinical trials evaluating the success of RT in combination with targeted therapy (18). The therapeutic effect of these drugs may be improved in tandem with radiation treatments, for example when the drug targeted molecules are even regulated by RT. This is the case of the combined use of RT and EGFR inhibitors, such as cetuximab. This molecule is indicated for patients with metastatic colorectal cancer, positive for epidermal growth factor receptor (EGFR) and without RAS mutations (wild-type) under mono-regimen or in association with chemotherapy. Together with RAS mutation, also BRAF mutation affects the efficacy of this monoclonal antibody; both these two genes are included in the EGFR transduction pathways.
Clinical studies have supported the approval and implementation of cetuximab in clinical treatment regimens of neck squamous cell carcinoma (HNSCC) in combination with RT with encouraging results (19,20). In patients with BRAF/KRAS wild-type rectal carcinoma, receiving neoadjuvant radio-chemotherapy, cetuximab increased overall survival and radiologic tumor response rate, while there was no significant effect in the whole patient population (including patients with both wild-type and mutated BRAF/KRAS tumors) (21).
Similarly to EGFR antibodies, EGFR tyrosine kinase inhibitors (erlotinib, gefitinib, afatinib) have been found to be more effective in a specific subset of patients, having mutations in the EGFR kinase domain (22,23). Later, several recent phase II studies describe promising results for the use of EGFR kinase inhibitors in combination with RT (24-26). In these trials, when erlotinib was added to the treatment regimen, the outcome and response in patients with advanced stage NSCLC were better than the results from published studies (24-26). Interestingly, Komaki et al. did not find a correlation between EGFR mutation status and response, although this may be due to the relatively small number of mutated patients included in the studies (24).
One other example of how mutations can affect the efficacy or the resistance to a certain treatment can be shown considering clinical studies which analyze the efficacy of BRAF inhibition in combination with RT. As it is known, BRAF mutation V600E is frequent in melanoma tumors and the inhibitors vemurafenib and dabrafenib are designed to inhibit only BRAF V600E, but not wild-type BRAF (27). Satzger et al. reported no response to the combined treatment of RT plus dabrafenib or vemurafenib in patients with metastatic melanoma. In addition, some patients experienced severe radiation dermatitis, indicating a radiosensitizing effect on normal tissue with wild-type BRAF, but not on malignant cells harboring BRAF V600E mutations (28). This further emphasizes how molecular targeted drugs can produce different effects when they are used in combination with radiation compared to monotherapeutic application, modifying the TCP/NTCP balance.
Other “omic” biomarkers of tumor profiling to be used in precision medicine in combination with targeted therapies are extensively reviewed in Eke et al. (18).
The concept that tumor mutations can affect RT results has been recently showed even for hadrontherapy. The group of Amornwichet (29) has investigated the association between the mutational status of EGFR and KRAS, driver genes frequently mutated in NSCLC, and the relative biological effectiveness (RBE) of carbon-ion beams over X-rays. In a selection of 15 NSCLCs having different KRAS/EGFR mutation patterns, the authors evidenced that EGFR-mutant NSCLC cells, but not KRAS-mutant cells, show low RBE. Then, NSCLC patient carriers of EGFR mutation might not benefit from carbon-ion radiotherapy (CIRT) (29).
Proteo-genomic approaches by using cell lines as in in vitro models, also contribute to highlight tumor response to radiation. Today, increasing evidences show that different rates of sensitivity to treatments are cell-type and dose-dependent (14,15,30). Our research group, by a proteogenomic study, observed different rates of cell survival and process activation (e.g., senescence induction) with differential molecular profiles at transcriptional and protein level, in tumorigenic and non-tumorigenic breast cancer (BC) cell lines (MCF7, MDA-MB-231, MCF10A) and in primary BC cells treated with high doses (9 and 23 Gy) of IR by electron beams (14,15,30). We described a panel of genes regulated by 9 and 23 Gy doses in the cell lines analyzed, and suggested a dose- and cell type- dependent transcription, able to affect cell fate modulating some cellular processes such as DNA repair, inflammation, cell death and cell cycle (14,15,31). These results suggest that tumor radiosensitivity is difficult to be predicted in advance, nonetheless the omic studies have revealed several biomarkers able to affect RT outcomes. Moreover, proteogenomic studies showed that each tumor type may activate different pathway addressing each own cell fate in specific manners (apoptosis, necrosis, senescence, autophagy, mitotic catastrophe) (32). This aspect should to be keep in count when RT is associated to other targeted therapies, as they could become an efficacious army against cancer if synergize on pathways activated by RT. One major factor is intra-tumoral heterogeneity, with a range of differences in the expression of RNA, proteins and metabolites among sub-populations within the tumor. Consequently, in the evaluation of prognostic molecular profiles, this heterogeneity within one tumor can obscure potential correlations with survival or disease progression.
One possibility to overcome the lack of predictability of RT outcomes, due to the complexity of tumor heterogeneity, could be the introduction of functional tests, based on clonogenic assays, on primary cells isolated from bioptic fragments to predict both tumor and normal tissue radiosensitivity (33). Then, the calculation of % of surviving fraction from in vitro treatment of primary cells, during the latency time from surgery to the beginning of RT treatment, could help to obtain measurable indications of tumor sensitivity, when no other biological molecular biomarkers of radiosensitivity are known, to be introduced in TCP modelling, permitting to connect the absorbed doses with clinical outcome. Alternatively, the survival fractions obtained in a dose-response curve, by in vitro treatment of primary cells, could permit to describe response to IR by linear quadratic-model or its adaptive models, which have been explored to overcome scenarios in which the α/β ratio fails to sufficiently reflect differences between dose-response curves (34).
Influence of CSCs
CSCs were first identified in human acute myeloid leukemia (AML) cells in 1994 by Lapidot et al. These authors isolated AML cells expressing specific cell surface markers and detected that a CD34+CD38- population were able to engraft severe combined immune-deficient (SCID) mice and develop progenitors of human leukemia. On the contrary, the CD34+CD38+ and CD34- cell fractions were not able to induce such responses (35). Despite the existence of CSCs in solid tumors has been early debated, continuous CSC characterization supports their existence. Indeed, CSCs were identified in many solid tumors, including breast, prostate, pancreas, brain, colon, liver, lung, ovary and skin cancers (36-38). The CSC theory suggests that a subpopulation of cells in the tumor detains stem cell properties with the potential to self-renew and generate the entire heterogeneous tumor bulk in a unique hierarchic pattern (39,40). In the hierarchic model CSCs are a distinct subpopulation of cells that drive both tumorigenesis and metastasis. These cells reside at the top of neoplastic hierarchies and divide symmetrically and asymmetrically in a similar pattern to normal stem cells (SC). In addition, the stochastic model has been proposed to demonstrate that a tumor develops as a result of random oncogenic mutations. Some authors believe that the hierarchic and stochastic models of cancer are not mutually exclusive, adding another level of complexity to understanding of cancer biology (41,42). To date, several evidences suggest that CSCs may be responsible for both chemo- and radioresistance, leading to cancer cell survival, invasion, metastasis and further recurrence (43-45). It has been ascertained that cancer cells are heterogeneous in their radiation response and CSCs are most resistant to radiation possessing specific molecular properties protecting it against radiation-induced damage (45,46). The degree of radiosensitivity is recognized to be related to both intrinsic properties, cell-type specific, including DNA repair, cell cycle status, survival/cell death pathways and extrinsic properties which include molecular signals from the extracellular microenvironment. It is assumed that these combinatorial factors enable CSCs to withstand radiation injury (47,48).
In order to develop useful targeted approaches to increase the CSCs response to RT, it is necessary to highlight the molecular features that contribute to CSC radiosensitivity/radioresistance. Understanding the signaling pathways that determine radioresistance is crucial for selecting appropriate treatment modalities for patients and developing novel molecular agents to enhance radiosensitivity in human cancers.
Cell cycle status and DNA repair
Cell cycle has an important role in radioresistance. The activation of cell cycle checkpoint kinases, Chk1 and Chk2, was found in CD133+ glioblastoma CSCs (GSCs) compared with CD133− non-CSCs (49). Zhou et al. showed that U87 and U251 glioblastoma SC were more radioresistant compared to glioma cells (GCs) due to high expression of phosphorylated cell cycle checkpoint proteins such as ATM, p53 and Chk2. ATM inhibition induced cell cycle checkpoint defects and increased the rate of apoptosis of GSCs following X-rays treatments with several doses (2–4–6–9 Gy). Therefore, ATM may represent a factor of radioresistance and a target of improved radiosensitivity in GSCs (50). For example, the CP466722 compound (Pfizer), an ATM kinase inhibitor was identified (51). CP466722 inhibited ATM-dependent phosphorylation events and the disruption of ATM function resulted in cell cycle checkpoint defects. The ATM kinase activity blockade was completely removed after withdrawal of CP466722, showing that short-term inhibition of ATM was sufficient to sensitize cells to radiation. Thus, drugs such as CP466722 provide important tools to stop constitutive activation of cell cycle checkpoints in CSCs of GCs and allow more sensitivity to radiotherapy (45,51).
An additional property of CSCs associated with radioresistance is their ability to remain in a quiescent state. This property makes them more resistant to cell cycle related agents, including many chemotherapeutic drugs, such as paclitaxel and radiation (52).
It is known that proliferating cells are more radiosensitive than quiescent cells as the cells in G2/M phase are most radiosensitive, while those in late S phase are most radioresistant (44,53). It has been demonstrated that during fractionated RT, the loss of the bulk tumor cells induces re-entry into the cell cycle and accelerates CSCs repopulation (44). Abnormal regulation of cyclin-dependent kinase (CDK) pathways controlling cell cycle progression, such as the p16-CDK4-RB pathway, may promote CSCs generation and proliferation (54,55) . Hence, it is crucial to determine the balance between triggering CSCs into cell cycle and uncontrolled proliferation during and after irradiation.
DNA repair is also involved in CSC-associated radioresistance. It was showed that enriched CSC cell populations such as CD44+/CD24− BC cells and CD133+ GCs displayed increased DNA repair capability compared with non-CSC-enriched cell populations (56,57).
Glioma CSCs play a crucial role in radioresistance through activation of DNA damage checkpoint proteins including ATM, SMC1, Chk1, Chk2, and p53 and increased DNA repair. It was reported that CSCs exhibited more efficient DNA damage repair than bulk tumor cells when exposed to radiation (47,58). Gene expression profiling studies by microarray revealed an increased DNA damage response and expression of DNA repair genes among BC CSC (59).
Signaling pathways and radioresistance
Different signaling pathways have been shown to be responsible for both CSC self-renewal and radioresistance in multiple cancers, some of these are specific and frequently involved, including the Notch, Hedgehog (Hh), and Wnt pathways (47) (Figure 2).
Notch pathway is a well-known CSC pathway. It is a highly conserved cell signaling system active in a wide range of human tumors promoting self-renewal and repressing differentiation (60-62). High Notch pathway activity showed correlation with poor prognosis and radioresistance in NSCLC patients (63). Inhibition of Notch pathway with gamma-secretase inhibitors (GSIs) rendered the glioma SC more sensitive to radiation at clinically relevant doses, suggesting a critical role of Notch signaling to regulate radioresistance of glioma SC (64). These and other evidences highly reveal that Notch signaling pathway increases the accelerated repopulation and radiation resistance properties of CSCs (44,65). However, the exact mechanism by which radioresistance is mediated by Notch signaling is not yet clear and needs further study.
The Hh pathway is thought to play an important role in regulating CSC proliferation, survival and maintenance (66,67). Many studies have demonstrated that overexpression of Hh signaling genes is linked to radiation resistance and downregulation can enhance radiation responses in many tumor types. In human GCs, Hh signaling regulates the expression of stemness genes and the self-renewal of CD133+ CSCs (68). Hh signaling pathway activation may promote the repopulation of CSCs after RT, contributing to both radiation resistance and treatment failure. Analysis of esophageal tumor samples from RT resistant cancer patients showed that 83.7% had activated Hh signaling defined by Hh expression and nuclear GLI transcription factor localization (69,70). Li et al. demonstrated that the Hh pathway played a key role in resistance to RT and that Hh pathway inhibition may sensitize AML cells to radiation by overcoming radioresistance (71).
Wnt/β-catenin pathway is another important CSC signaling pathway whose activation has been shown to maintain CSC self-renewal in many different ways, including enhancing CSCs proliferation status and controlling CSCs capability to be associated with their niches (61,72).
This pathway was reported to be associated with cancer metastasis in breast, lung and prostate cancers (73), and it was also found to be involved in CSC radioresistance (47). Woodward et al. showed that the CSCs subpopulation was enriched after radiation in mouse models of breast cancer and the level of activated β-catenin was elevated, suggesting a role for the Wnt/β-catenin signalling cascade and revealing a more effective DNA repair in CSCs after radiation (74). Chang et al. elucidated the mechanism of cell death after radiation by studying how β-catenin silencing controls the radiation sensitivity of radioresistant head and neck cancer cells. The authors showed that Wnt/β-catenin silencing led to a significant reduction in cancer cell radioresistance associated with a decreased activity of Ku70/Ku80 DNA repair machinery (75). Although the role of Wnt/β-catenin signaling in radioresistance of CSCs remains not entirely clear, Wnt signaling activates DNA damage response and leads to tolerance of DNA damage and resistance to radiation. It is hypothesized that this genomic instability may drive the transformation of normal SC into CSCs (76).
Influence of hypoxia
It has long been known that oxygen concentration below normal physiological levels is often found in tumors and it may influence malignant progression. Indeed, human tumors contain regions of acute and chronic hypoxia associated with poor prognosis and metastatic disease. Chronic hypoxia represents the more typical situation in tumors because of an insufficient vascularization and limited diffusion of oxygen into the tissues (77,78). Cancer cells that adapt to the selective pressure of hypoxia and acquire a “hypoxic phenotype” tend to form more aggressive and invasive tumors that are resistant to traditional therapies. Prolonged and reduced oxygen concentration strongly influences radiation response. Long since many evidences demonstrate that oxygen is a potent radiosensitizer which can increase the effectiveness of radiation and that hypoxic cells are 2–3 times more resistant to radiation (79,80). Under normoxic conditions IR generates ROS (Reactive Oxygen Species) and free radicals through a direct and indirect effect that cause irreparable DNA damage and cell death. Oxygen is able to react with free radicals to yield a stable change in the chemical composition of the DNA damage. Intratumoral hypoxia hampers ROS formation, interferes with the fixation of DNA damage resulting in inefficient DNA strand breaks and radioresistance (81). A major concept in clinical radiobiology is that tumor subpopulations in hypoxic areas are critical to target therapeutic effects and reoxygenation between dose fractions is generally believed to improve the efficacy of radiation treatment by increasing tumor radiosensitivity (43). The length of time that cells are under hypoxic conditions and the extent of the hypoxia are critical factors in terms of the biological cell response to radiation. The underlying mechanisms are not entirely known but it is suggested that the radioprotective effect of hypoxia does not exclusively rely on the radiochemistry of oxygen but rather involves complex signaling events, mediated by transduction pathways, with homeostatic effect if the hypoxia persists long enough, thereby losing its protective effects (77-79).
The cellular response to hypoxia is mainly controlled by the hypoxia-inducible factor (HIF) family of transcription factors, primarily mediated by the HIF-1 and HIF-2, which regulate the expression of multiple genes involved in processes that lead the adaptation and progression of cancer cells, including cell proliferation, metabolism, immune responses, genomic instability, vascularization, invasion and metastasis. HIFs contribute to chemo-and RT resistance through many mechanisms, and in tumor samples HIF expression is often associated with poor prognoses and relapse on treatments (82,83). HIF-1α expression is transiently decreased after IR treatments owing to re-oxygenation of surviving hypoxic tumor cells. ROS production next stabilizes the HIF-1α protein, increasing HIF-1α expression and its target genes (82-84). HIF-1α exerts both radiosensitizing effects, by promoting proliferation and apoptosis often p53-induced, and radioresistant effects by vascular radioprotection. Activation of the HIF-1 pathway represents a cellular defense mechanism against irradiation. Following to HIF-1 upregulation after RT, tumor cells secrete VEGF and CXCL12, which stimulate angiogenesis and vasculogenesis, resulting in recovery of tumor blood, nutrient supply and tumor recurrence. Alterations in tumor glucose metabolism, HIF-1α-mediated, may also affect cellular responses to IR (83,84). An active HIF-1 pathway also leads to changes in tumor glucose metabolism with accumulation of lactate, pyruvate, gluthathione, and NADPH. These molecules remove free radicals and ROS, protecting the tumor cell from free radical–mediated DNA damage. Furthermore, lactate stimulates endothelial cells to produce VEGF, thereby contributing to angiogenesis (85).
Genes that are up- or downregulated in response to hypoxia reflect the hypoxic phenotype and can provide an indirect measure of the hypoxia level. Several studies performed gene expression profiling by microarrays to highlight a global transcriptional response to hypoxia or by using an in vitro approach with cell lines exposed to hypoxia versus normoxia or analyzing clinical samples. Genes found to be significantly upregulated, or exceeding a defined threshold from baseline normoxic expression, were grouped together and referred to as a “hypoxia gene expression signature” or “hypoxia signature”. The evaluation of individual genes or common pathways in hypoxia signatures contributes to identify potential new therapeutic targets (86).
Considering hypoxia as a hallmark of solid tumors that mediates metastatic and treatment resistant neoplasia, it represents one of the most attractive therapeutic targets in cancer. Several approaches for targeting hypoxic tumor cells have been proposed including hypoxia-activated prodrugs, gene therapy, specific targeting of HIFs, or targeting of relevant pathways in hypoxic cells such as the mTOR and UPR (unfolded protein response) pathway (83). Targeting HIF-1 and tumor glucose metabolism at several levels reduces the antioxidant capacity of tumors, affects the tumor microenvironment, and sensitizes solid tumors to irradiation (87,88). Silencing or pharmacological inhibition of HIF-1α can increase the anti-tumor effects of IR. Indeed, HIF inhibitors can be used in combination with RT to target RT-resistant tumor cells. Furthermore, radiosensitizers such as misonidazole mimic the effect of oxygen and can be used to enhance IR efficiency in hypoxic tumors, particularly in head and neck cancers (87-89).
In addition, several evidences show the crucial role of CSCs and oxygen on tumor radioresistance (Figure 2). A state of hypoxia in the niches is necessary to maintain CSCs in an undifferentiated state that positively regulate the expression of CSC surface markers (e.g., CD133 and CD44), and transcription factors, such as SOX2. CSC survival can be regulated by their niches following radiation exposure (90-92). Indeed, upon irradiation, direct interactions between CSCs and their surrounding cells have been shown to upregulate some anti-apoptotic proteins, like Survivin and BCL-2, and decrease the apoptotic response in prostate cancer cells (44). Furthermore, knockdown of Survivin by antisense oligonucleotides significantly sensitizes the radiation response of colorectal cancer cells both in vitro and in vivo (93). CD133+ glioma SC express higher levels of protective autophagy proteins after irradiation, and resistance can be attenuated by inhibition of the expression of these proteins in vitro sphere-forming assays (94,95). In addition, niche-associated Notch pathway may be activated after irradiation with increased symmetric cell division and accelerated repopulation of CSCs. The Notch pathway also plays a crucial role in linking angiogenesis and CSC self-renewal in glioblastoma CSCs. Notch induction may activate some other pathways, such as the EGFR pathway, which could promote DNA repair capability and CSC survival (96-98) . Moreover, CSC niches may also produce survival cytokines, such as EGF, FGF, and VEGF, all of which affect cancer cell radioresistance and radioprotection (44,91,92).
In association with hypoxia, HIF-1 expression is increased in CSCs, which may be protected from oxidative damage with augmented ability of DNA damage response and resistance to cell death induced by radiotherapy (99,100). In addition, TGF-β, a stem cell related pathway, may induce HIF-1 stabilization confirming the importance of CSCs to reside in a hypoxic environment by interacting with their niches (44,99,100).
Genetics and “omic” biomarkers of radiation toxicity
Clinical end points
Adverse effects to RT could be described as early and late reactions. All patients will experience toxicity with variable grading from minor to severe effects, and duration from weeks to lifetime.
The early effects mainly involve tissues with rapid turnover, such as epithelium and immune cells. The tumor control is closely related to clonogenic inactivation of tumor cells by dose escalation, thus treatments could be very detrimental for tissues as epithelia, which are characterised by continuous cell renewal with a stem cell niche and a transit-amplifying cell (TAC) compartment. In normal tissue homeostasis, SC divide asymmetrically producing a TAC progenitor and a stem cell remaining in the niche. This mechanism is finely preserved and regulated by shedding or apoptosis of terminally differentiated cells (101,102).
For this reason, crypt and villi of the small intestine are particular sensitive to radiation, as a niche of 4–6 cells is protected in the crypts, which produce TACs. The loss of one stem cell, will drive to a temporary symmetric division, replacing the staminal reserve. However, if all the SC are lost, the villus will disappear within days (103).
For “n” SC in a crypt, the fraction of inactivated cells after a dose D resulting in a surviving fraction, SF, will be (1-SFD)n. Thus, a dose resulting in SFD =0.01 will inactivate 4–6 SC with 94–96% probability. From radiation accidents and experimental animal studies doses D ≥10 Gy to the small intestine are known to be lethal (33).
A similar behaviour has been demonstrated for the SC reserve of mouse tongue epithelium. In this case, SC are slowly depleted during the early part of a fractionated schedule, driving to a symmetric cell division in order to replace the rate of cell loss during the remaining part of the fractionated schedule (104,105).
Acute toxicity involving proliferating tissue, usually affects the skin with the occurrence of erythema, dermatitis, desquamation and hair loss, or intestine causing diarrhea or bladder causing cystitis (106). However, these problems are generally transient.
Moreover, immune reactions also play an important role in local and systemic response to radiation treatments, towards the involvement of mediators as inflammatory cytokines, growth factors and proteases which can affect cancer cell invasion, bystander effect, radiation tissue complications such as fibrosis, genomic instability and thus can greatly affect intrinsic cellular radiosensitivity and side effects occurrence (31). In vitro experiments show that radiation treatments induces the release, within the first 72 h, of cytokines and growth factors potentially able to affect the tumor outcome (progression, invasiveness, cell survival balance, bystander effects) with a dose-independent and cell-line dependent signature (30). Inflammation has the consequence of producing ROS species, which produce further oxidative DNA damage even in normal tissues surrounding the irradiated ones. Then, inflammation post radiation produces early and late toxicity effects. Late effects mainly regard fibrosis and senescence induction. Although irradiated fibroblasts do not undergo rapidly to apoptosis and are referred to be senescent, their better profile definition is “terminal differentiated”. Indeed they remain in metabolically active for months or years, even if permanently arrested, releasing a great quantity of matrix products whereas their proteolytic power is reduced by the downregulation of metalloproteases (MMPs) and their inhibitors upregulation (107-109).
Fibrosis could have effects impacting on life’s quality, such as producing bowel malabsorption or bowel and urethra obstruction and then incontinence, after pelvis irradiation. Moreover, vascular damage is responsible of telangiectasia, which is visible on skin as small vessel dilatation, or even hematuria. Conversely, vascular damage can cause vessels constriction, causing ischemia and necrosis, resulting in vessel perforation and fistulae. Moreover, among radiation side effects, it can be remembered hormone deficiency and infertility.
In addition, previous studies on RT-induced skin telangiectasia suggested that up to 80% of the observed variation in risk was associated with individual patient-related factors, after considering the effects of absorbed dose and dose per fraction (110,111). Indeed, among normal tissue complications, neither erythema nor subdermal fibrosis correlated with telangiectasia of the skin in BC RT patients (8). However, a more recent larger study showed a significantly increased risk of fibrosis in breast RT patients with telangiectasia, although overall, the risk of developing fibrosis was much lower than that of telangiectasia (112). Conversely, the RAPPER study (“Radiogenomics: Assessment of Polymorphisms for Predicting the Effects of Radiotherapy”), which comprises 778 breast RT patients enrolled in the Cambridge breast IMRT (intensity-modulated RT) trial, have shown a lack of correlation radiation-induced fibrosis and telangiectasia, nonetheless significant correlations were found between several end points. This supports the view that there are differences in the mechanisms underlying the pathogenesis of these two end points (113).
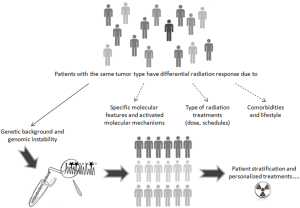
In addition to the above described deterministic side toxicity effects, other stochastics events such as genetic mutations represent a cause of radiation induced second tumors, which is appearing among long surviving patients with the success improving of cancer care. The relative risk to develop a second cancers after RT in adult patients is estimated in 1.2 in recent meta-analyses, even larger in children (114). In this context, the genomic instability is considered to be an important risk factor (115,116). These events are generally not predictable and individual radiosensitivity became the results of interaction between internal (complex genetic background, different molecular mechanisms, cells, and pathways) and external factors (dose or dose per fraction). Other risk factors are comorbidities, concurrent treatment, smoking and lifestyles (Figure 3). Then, functional test predicting individual sensitivity are generally not applicable, although individual variability can count for about 80% of normal tissue radiosensitivity (110). However, some particular mutations underlie to specific tissue or body radiosentitivity (ATM). These aspects are detailed in the paragraph below.
Genetics of radiosensitivity
The 1990s have marked the beginning of the omic science era, that revealed us a new vision on the individual susceptibility to develop diseases, to reach longevity and even in conditioning the RT success. It became clear that only few mutations are responsible of extremely high normal tissues radiosensitivity, whereas a complex genetic background of single nucleotide polymorphisms (SNPs) throughout the entire genome underlie of a huge variability of response to treatments (Figure 3).
Patients suffering from certain rare genetic syndromes such as ataxia telangiectasia, Blooms syndrome, Fanconi’s anaemia and Nijmegen breakage syndrome experience devastating normal tissue reactions if treated with radiotherapy (117). All these syndromes are related to mutations in genes involved in detection of DNA damage and initiation of DNA repair. However, there are no evidences that heterozygous carriers of ATM, BCRA1 and BCRA2 genes manifest strong normal tissue complications post- radiation treatment. Similarly, no obvious association was found between clinical normal tissue radiosensitivity and mutations in other DNA repair genes like RAD50, RAD21, NBN or MRE11A (118).
A comprehensive review well describes candidate pathways having a key role in the cell fate balance after treatment with ionizing radiation (17).
According to a recent review, a total of 128 studies have investigated genetics of normal tissue radiosensitivity (119). However, the majority of these studies are small, with a median of cases under 150 patients, whereas only ten studies include more than 500 cases. Then, most of the studies lack to detect the small effect sizes usually featuring SNPs and few associations are replicated in independent studies. So, most of results are inconsistent and only few SNPs could be addressed as genetic markers of normal tissue sensitivity. SNPs more often investigated are sited in genes involved in DNA damage detection (ATM), DNA repair (XRCC1, XRCC3, APEX), tissue remodelling (TGFB1 and TIMP) and scavenging of reactive oxygen species (SOD2 and GSTP1), which represent main mechanisms regulating radiation induced response. Many other works have evaluated the role of specific candidate genes (TGFB1, XRCC3, XRCC1, ATM, GSTP1, GSTA1, TP53) in radiotherapy induced toxicity in breast and gynaecological cancer cohorts, with contrasting results (33).
The candidate gene approach has been considered to be unsuccessful, then larger collaborative radiogenomics studies, involving few thousands of patients, have been proposed, some of which finding replicated associations (120-122). In 2010, the Radiogenomics Consortium (RGC) was established in order to collect patients cohorts and resources for large-scale GWAS on normal-tissue radiosensitivity (123,124).
Regarding the TGF-β signaling pathway, a study has evaluated the role of 43 SNPs in 35 genes belonging to TGF-β pathway in 2,036 patients in three independent cohorts, showing an increased risk of late toxicity [OR 2.45, 95% confidence interval (CI): 1.52–3.98] for patients being homozygous for the minor allele of the rs1800629 SNP. Its exact functional role is unknown, although it is sited a few hundred bases upstream of the transcription start site of the TNF-α gene, relatively close to the lymphotoxin alpha gene (LTA), and close linkage with a SNP in a MHC class III (120). Similarly, a study designed in two phases, preliminarily tested 305 SNPs in 59 genes related to oxidative stress and DNA repair pathway for their role in inducing side toxicity in 753 breast cancer patients. Then, the top ranking 10 SNPs were replicated in independent cohorts including a total of 1883 patients. Using this approach, the SNP (rs2682585) sited in the DNA repair gene XRCC1 was identified as a protective marker of toxicity, as carriers of the minor allele had a reduced risk of late skin toxicity (multivariate OR 0.77, 95% CI: 0.61–0.96, P=0.02) and overall toxicity (regression coefficient −0.08, 95% CI: −0.15 to −0.02, P=0.02) (122).
Furthermore, in a recent study of 2014, the ATM rs1801516 SNP was identified as risk factor of toxicity in a large study of 5,456 breast and prostate cancer patients cohort, with an OR of approximately 1.5 for acute toxicity and 1.2 for late toxicity (125). This study makes evident the dependence of side effects occurrence by the individual genetic background and puts the attention on the population-dependent allele frequency and even on the tissue-dependent gene activation, which control a different gene transcription level in tissue and organs. These differences increase the variability in tissue toxicity in cohort of patients from different geographic area. To make an idea of geographic impact on genetics, it can mention the Italy example, where Sicilian individuals could be considered genetically different from Italians with a south-north trend, and Sardinians are more rich of centenarians respect to the rest of Europe (126,127).
Genome wide association studies (GWAS) started with the idea of “fishing in the ocean”, in order to discover complex genetic profiles responsible of a certain phenotype, constituted by multiple allele variation with low penetrance and small size effects. This comparison is due to the fact that SNPs with strong associations are located close to the sea’surface whereas very large cohorts of cases are needed to find weak associations (119).
One large GWAS study has been conducted on a total of 3588 breast and prostate cancer cohorts, using the STAT score to provide evidence that genetic polymorphisms are associated with late toxicity. However, this study evidenced the fact that stronger associations could be detected referring to comparisons between specific end point for specific treated tissues. Then, breast and prostate cancer patients suffer of different side effects affected by different risk’ alleles (128). The same group identified the locus comprising TANC1 at 2q24.1 as late toxicity risk factor in a three-stage genome-wide association study, performed on three prostate cancer cohorts from different geographic area (Spain, UK and North America), consisting of 1,742 patients. The inclusion of the third cohort gave un adjusted Pcombined=4.64×10-11 (129).
A very recent GWAS has combined the results from four radiotherapy cohorts: RAPPER (n=533), RADIOGEN (n=597), GenePARE (n=290) and CCI (n=150), investigating whether prostate cancer patients with a high genetic risk have increased toxicity following radiotherapy. Patients with a high polygenic susceptibility for prostate cancer have no increased risk for developing late radiotherapy toxicity. These findings suggest that patients with a genetic predisposition for prostate cancer, inferred by common variants, can be safely treated using current standard radiotherapy regimens (130). Other few GWAS studies were conducted in the past years, often considering prostate cancer treated cohorts, which are very sensitive in the development of radiotherapy induced early and late toxicity.
The first one was published in 2010 (131), involving 79 prostate cancer treated patients African Americans and investigating the risk of erectile dysfunction. It identified a SNP (rs2268363) located on the gene of the follicle-stimulating hormone receptor (FSHR) with an OR of 7.03 and a P value of 5.46×10−8. On the other hand, another two stage GWAS study conducted in 2013, evaluating again the erectile dysfunction’ risk on a total of 465 failed to find significant associated SNPs under the usual genome wide significance level of 5×10−8 (132). Nevertheless, the study identified a SNP (rs11648233) close to the 17-betahydroxysteroid dehydrogenase II gene (HSD17B2), that catalyses the oxidative metabolism of androgens and estrogens (OR 1.8; P value of 9.1×10−5).
Additionally, again on prostate cancer cohorts, other two two-stages designed GWASs were published in 2013. The first one included 723 patients (133) and showed a borderline significant association between a haplotype of 8 SNPs encompassing the interferon kappa (INFK) gene and overall urinary toxicity (OR 2.5 and; P value of 6.5×10−7). The second one included 1,149 prostate cancer patients (134) and reported an association between two SNPs in ‘a gene desert’ (rs7120482 and rs17630638) and the risk of rectal bleeding after radiotherapy (OR 6.7 and 3.1 in first and second stages respectively; P valuecombined =5.4×10−8).
The studies described herein reveal a poor results repeatability, nonetheless the efforts to design GWAS adequately powered also in the contest of collaborative research projects within international consortia like the RGC. The reasons are several. The identified SNPs have very small impact on phenotype, whereas the SNPs having a major role are rare (e.g., ATM, BRCA1, BRCA2, etc.). One more reason is due to the fact that SNPs association is searched with different end points among studies, involving different tissues, so that results become not repetible. Furthermore, but not less important, it have to be hypothesized that individuals carriers of risk SNPs may also be carriers of some other protective SNPs or non-coding SNPs affecting differently gene expression in different tissues.
Moreover, mitochondria DNA (mtDNA) can also participate to affect the balance in radiosensitivity and the occurrence of late reactions, thanks to the presence of genes involved in controlling ROS metabolism. Alsbeih et al. performed a case-control study in order to test the hypothesis that mtDNA genetic variations can contribute to patient-to-patient variability in normal tissue response to RT with encouraging results (135). In particular, a significant association (P=0.01) was observed for the non-synonymous A to G substitution at nucleotide 10398 located in the respiratory gene NADH dehydrogenase subunit 3 (ND3), leading to a change in the amino acid sequence from Thr to Ala at codon 114. The G-variant was more frequent in the radiosensitive group than in the control group (odds ratio, 7.2; 95% CI, 1.16–51.65) (135).
Individuals having a balanced profile do not experiment toxicity and/or those patients are not included among patients with toxicity. Functional test of predictive assays may help to identify patients as being radiosensitive for one adverse event while remaining radioresistant to another (136). Then, the success of individual radiotherapy depends from complex profiles, nonetheless bio-informatic power and genotyping technology advances provide great opportunities to identify genetic biomarkers of normal tissue radiosensitivity.
Omics of radiosensitivity
A recent novelty to improve the discover of allele association risk of normal tissue radiosensitivity consists in studying genes designated as ‘expression quantitative trait loci’ (eQTLs), which are variants affecting gene expression levels (137). These are classified in “local and distant eQTLs”, if the distance between the variant and the gene which it regulates is respectively below or above 1–2 mega bases (Mb). In the second case, the variant can regulate the expression of gene located on different chromosomes. Obviously, the eQTLs affect gene expression in a tissue-, cell type- and stimulus- specific mode and then, eQTLs may be associated with various genes, even if identified eQTLs are enriched of associated SNPs detected in GWASs and vice versa (137,138). One large NIH-funded project, known as the Genotype Tissue Expression (GTEx) project, will map eQTLs in 60 different tissues from 900 individuals (139).
The experimental scheme to address normal tissue radiosensitivity could be to culture in vitro fibroblasts from a certain number of patients that undergo to RT treatment. Fibroblasts could be exposed to radiation and gene expression should be quantified before and after irradiation. GWAS could be performed on recruited patients. Then, rad-eQLTs could be identified merging GWAS and gene expression data and, thus, tested for a possible association with risk of radiation induced fibrosis in a greater cohort of patients.
Few studies have addressed eQTLs after in vitro irradiation. The first one was published in 2009 and involved subjects from 15 families. In vitro irradiation was conducted on immortalized B-lymphocytes and the analysis identified around 1,200 loci associated with the transcriptional response to irradiation at a significance level of 4×10−5 (140). Moreover, some associated rad-eQTLs were confirmed by functional tests. Other two studies show associated rad- local and distant eQTLs, using a similar approach (121,141).
A discrete number of studies searched biomarkers of normal tissue radiosensitivity using other “omic” technological approaches, some of theme summarized in Table 1. A recent review resumes in a table the list of publications regarding “omic” studies on RT toxicity (33). The majority could be considered single experiments, so that no other useful omic biomarkers could be considered at a good level of validation to be associated to normal tissue complication.

Full table
Transcriptomic studies are usually conducted exposing cell cultures derived from patients to IR such as lymphocytes, fibroblasts or skin culture. In a work of 2004, the group of Rieger exposed lymphoblastoid cells derived from 14 patients with acute radiation toxicity to ionizing and UV radiation. Because skin cancer is associated with UV radiation exposure, skin cancer patients were included as controls to ensure that they would not be assigned a high risk for radiation. They found a 24 gene signature which predicted radiation toxicity in 9 of 14 patients with no false positives among 43 controls (P=2.2×10-7), including genes involved in DNA repair, stress response, protein degradation, and apoptosis (142).
Then, Svensson and colleagues, analyzed the severe late radiation toxicity of 21 prostate cancer patients that were compared with 17 patients without symptoms. The 24-h post radiation with 2-Gy X-rays exposure was analyzed by gene expression profiling and used for classification. A 72 gene signature, including genes involved in protein turnover, development, stress signaling and apoptosis, was able to discriminate cancer patients with and without late radiation toxicity (143). Other studies have evaluated the association from lymphocyte irradiated gene expression profile and the radiation induced early and late toxicity (144-147).
Without using an omic approach, a very recent publication by Koerdt et al., have evaluated the expression profiles of MAPK, NOS1, NOS3, and PIK3CA genes, by RT-PCR, in specimens from head and neck cancer patients with and without a history of RT, administered 6 months before surgery (148). Clinical data on the occurrence of cervical wound healing disorders (WHDs) were available. Expression analysis of patients with post-operative WHDs revealed a significant increase in MAPK expression compared to the control group showing no postoperative WHDs. PIK3CA showed a significantly increased expression in patients with a history of RT (148).
Many other efforts were dedicated to search for fibrosis gene expression signature post RT. One large study, involving 254 women treated with breast conserving therapy, 31 of whom developed fibrosis 3–7 years after RT, identified an 87 differentially expressed genes, involving in transcription, development and intracellular transport and localisation (149). Among these, the up-regulation of SERPINE1, coding for plasminogen activator inhibitor-1, PAI-1, was found confirming the increased protein levels observed in fibrotic conditions (150). Pathway analysis identified the TGF-β1 signalling and interleukin-2 pathways as the two major gene sets (149).
Moreover, as proposed by Rødningen et al., a 18-gene predictive signature for subcutaneous fibrosis after post-mastectomy RT was found to be able to discriminate among 31 patients with none/mild or moderate/severe fibrosis. This signature was identified after irradiation of fibroblast cultures with 3×3.5 Gy separated by 24 h (151-153). Then, the same group validated these finding on an independent cohort of 160 head-and-neck cancer patients of which 46 developed severe (grade 3) fibrosis. In agreement with the initial findings, the gene expression signature, using 9 of the 18 genes, was able to identify a smaller subset of patients that seems to be rather radioresistant and could therefore potentially be considered for dose escalation, whereas within the subset of patients with the ‘sensitive profile’, the cumulative risk of severe fibrosis was 34% at 9 years (Kaplan–Meier), and no patients with the ‘resistant profile’ developed severe fibrosis (P=0.035) (154).
Concerning the studies dedicated to asses proteomic biomarkers of radiation toxicity, a great attention has been dedicated to the role of circulating cytokines and immunological factors.
In particular, numerous studies have evaluated the role of TGF-β1 in tissue fibrosis. Some of these found higher circulating plasma levels of TGF-β1Jeny (155-157) in patients with radiation-induced late lung complications, while other studies found no correlation (158-160). According another one, the combination of elevated TGF-β1, lower IL-8, and mean lung dose is predictive for lung toxicity (155). No association was found between TGF-β1 and late toxicity in cervix cancer patients (161). Furthermore, A2M (alpha-2- macroglobulin) (162), C4BPA (complement component 4 binding protein, alpha chain), and VTN (vitronectin) have been recognized as novel biomarkers for RT-induced lung toxicity by using unbiased proteomic approach (163).
A minor attention has been dedicated to metabolomic studies performed to search for metabolic biomarkers of radiation toxicity, although a review discusses the role of the redox-sensitive metabolitetetrahydrobiopterin (BH4) in the pathogenesis of post-irradiation normal tissue injury, as well as how the metabolomic readout of BH4 metabolism fits in the overall picture of disrupted oxidative metabolism following IR exposure (164).
Another recent interesting preclinical work revealed that the urine metabolomic profile could predict the occurrence of a radio-induced neoplasm, although the latency time before a mass become detectable is very long. In this study, mice were exposed to 0 or 5.4 Gy total body irradiation (TBI), collecting urine samples periodically over 1 year. The analysis of urine metabolites by mass spectrometry revealed that cancers, including hematopoietic, solid, and benign neoplasms, could be distinguished and temporarily defined as early 3 months post-TBI for hematopoietic neoplasms, 6 months for solid neoplasms, and by 1 year for benign neoplasm, whewereas the metabolic signature 6 months post-TBI was found to be similar to non-irradiated control mice at 18 months, suggesting that TBI accelerates aging (165).
A comprehensive description of clinical and preclinical studies using metabolic signature to predict in advance radiation induced toxicity of organs and tissues, resulting from exposures to therapeutic or non-therapeutic IR, is reviewed in Menon et al. 2016 (166).
Finally, epigenetic modifications were even investigated to search for susceptibility bio-markers predictors of radiation induced toxicity.
Three serum miRNAs (miR-155, miR-221 and miR-21), related to immunity or inflammation were tested for their association with the occurrence radiation-induced esophageal toxicity (RIET) in non-small cell lung cancer (NSCLC) patients treated with chemoradiation therapy (CRT). The results showed that patients with stage IIIB-IV disease, higher mean esophagus dose or esophageal V50 had higher rates of severe RIET. In addition, high levels of miR-155 and miR-221 at week 1-2 of CRT were also risk factors for severe RIET (miR-155: OR 1.53, 95% CI: 1.04–2.25, P=0.03; miR-221: OR 2.07, 95% CI: 1.17–3.64, P=0.012), and the fold change of miR-221 was also predictive of severe RIET (OR 1.18, 95% CI: 1.02–1.37, P=0.026) (167).
A recent paper illustrates an epigenetic study on radiation induced fibrosis conducted on dermal fibroblasts obtained from BC patients before irradiation (168). A DNA methylation profiling identified a differentially methylated enhancer of diacylglycerol kinase alpha (DGKA). The authors deeply explored the biochemical consequences of different levels of transcription of this gene. In particular, they found that the DGKA inhibition has protective effects on diacylglycerol-mediated lipid homeostasis and reduces profibrotic fibroblast activation. Conversely, a decreased DNA methylation at this enhancer enables the recruitment of the profibrotic transcription factor early growth response 1 (EGR1) and facilitates radiation-induced DGKA transcription in cells from patients later developing fibrosis. These results suggest this differently methylated gene as marker of RT response. The same group reviewed this issue and listed nine hypermethylated genes and three hypomethylated genes as epigenetics markers of radiation-induced fibrosis (169).
Regards late side effects produced by chemo-radiotherapy treatment in cervical cancer patients, which count about 10–15% of treated individuals, an epigenetic study observed a significant association between promoter hypermethylation in the XRCC2 gene and occurrence of late grade III–IV toxicity (P=0.0357), among 18 candidate genes crucial in DNA repair and cell cycle regulation. This finding could be useful in the late toxicity prediction in RT-treated patients (170).
In addition, epigenetic biomarkers were searched in a preclinical study on radiation-induced skin fibrosis. In this case, a microarray analysis revealed significant epigenetic alterations for the miR-15a, miR-21, miR-30a and miR-34a. The last one was confirmed by other methods and their role was better explored. Particularly, the upregulation of miR-34a in radio-induced fibrosis causes inhibition of c-Met production, a known effector of fibrosis (171).
The combination between miR-99a higher levels and Ku 80 protein lower level in circulating lymphocytes were significantly associated (P=0.011 and P=0.013, respectively) with late rectal bleeding in prostate RT patients (172).
Finally, it has to be noted that, although histones modifications with the specific phosphorylation of Tyr-139 and dephosphorylation of Tyr-142 in histone H2AX are well established as immediate radio-induced effects, to our knowledge, no study have evaluated, still now, high-throughput screening of histone modifications in connection with normal-tissue reaction after RT.
Conclusions
Technological advances in radiation delivery to target tumors and the introduction of particle therapies have strongly limited the amount of dose distributed to normal tissues, enhancing the tumor killing capacity. Currently, RT treatment planning take in account physic metrics, nonetheless it emerges the necessity to include biological biomarkers of normal tissues and tumor radiosensitivity, to optimize therapeutic efficacy and prescribe an increased total dose to the tumor in patients with relatively radioresistant normal tissues and to address candidate patients having high risk of developing severe normal-tissue reaction to either different RT regimens.
However, patient selection based on a biological-driven treatment planning is still so far, as the RT final effect is complexly regulated by a balance between protective and risks biomarkers. Moreover, tumor heterogeneity represents an advanced level of complexity.
Our opinion is that functional tests on primary cells from patient biopsies by clonogenic assays, or imaging analysis, performed during the treatment scheme, could represent a modality to estimate tumor sensitivity to radiation. In addition, the evaluation of CSCs percentage and hypoxia area, need also to be evaluated in the modelling of tumor sensitivity. Finally, up to date only few significant gene alterations need to be taken in account in the modelling of normal tissue complication.
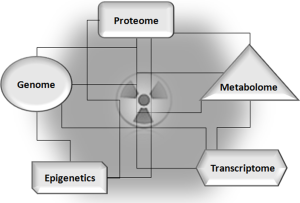
“Omics” biomarkers are, of course, needed to be identified and evaluated for their potential use to address combined treatments with targeted therapies able to increase radiosensitivity (Figure 4).
Acknowledgments
Funding: This work was supported by the INFN CSN5 Project Proposals: “ETHICS” (Pre-clinical Experimental and THeoretical studies to Improve treatment and protection by Charged particleS) and “MoVe IT” (Modeling and Verification for Ion beam Treatment planning).
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Translational Cancer Research for the series “Radiobiological models towards a personalized radiation oncology”. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.06.47). The series “Radiobiological models towards a personalized radiation oncology” was commissioned by the editorial office without any funding or sponsorship. GIF served as the unpaid Guest Editor of the series and serves as an unpaid editorial board member of Translational Cancer Research. GR served as the guest editors of the series. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kutcher GJ, Leibel SA, Mohan R, et al. Advances in precision treatment: some aspects of 3D conformal radiation therapy. Front Radiat Ther Oncol 1993;27:209-26. [Crossref] [PubMed]
- Elith C, Dempsey SE, Findlay N, et al. An Introduction to the Intensity-modulated Radiation Therapy (IMRT) Techniques, Tomotherapy, and VMAT. J Med Imaging Radiat Sci 2011;42:37-43. [Crossref]
- Mackie TR, Holmes T, Swerdloff S, et al. Tomotherapy: a new concept for the delivery of dynamic conformal radiotherapy. Med Phys 1993;20:1709-19. [Crossref] [PubMed]
- Orecchia R, Zurlo A, Loasses A, et al. Particle Beam Therapy (Hadrontherapy): Basis for Interest and Clinical Experience. European Journal of Cancer 1998;34:459-68. [Crossref] [PubMed]
- Kooy HM, Grassberger C. Intensity modulated proton therapy. Br J Radiol 2015;88:20150195 [Crossref] [PubMed]
- Marks LB, Ten Haken RK, Martel MK. Guest editor’s introduction to QUANTEC: a users guide. Int J Radiat Oncol Biol Phys 2010;76:S1-2. [Crossref] [PubMed]
- Emami B, Lyman J, Brown A, et al. Tolerance of normal tissue to therapeutic irradiation. Int J Radiat Oncol Biol Phys 1991;21:109-22. [Crossref] [PubMed]
- Bentzen SM, Constine LS, Deasy JO, et al. Quantitative analyses of normal tissue effects in the clinic (QUANTEC): an introduction to the scientific issues. Int J Radiat Oncol Biol Phys 2010;76:S3-9. [Crossref] [PubMed]
- Kutcher GJ, Burman C, Brewster L, et al. Histogram reduction method for calculating complication probabilities for three-dimensional treatment planning evaluations. Int J Radiat Oncol Biol Phys 1991;21:137-46. [Crossref] [PubMed]
- Baumann M, Krause M, Overgaard J, et al. Radiation oncology in the era of precision medicine. Nat Rev Cancer 2016;16:234-49. [Crossref] [PubMed]
- Bravatà V, Cammarata FP, Forte GI, et al. "Omics" of HER2-positive breast cancer. OMICS 2013;17:119-29. [Crossref] [PubMed]
- Minafra L, Norata R, Bravatà V, et al. Unmasking epithelial-mesenchymal transition in a breast cancer primary culture: a study report. BMC Res Notes 2012;5:343. [Crossref] [PubMed]
- Minafra L, Bravatà V, Forte GI, et al. Gene expression profiling of epithelial-mesenchymal transition in primary breast cancer cell culture. Anticancer Res 2014;34:2173-83. [PubMed]
- Minafra L, Bravatà V, Russo G, et al. Gene Expression Profiling of MCF10A Breast Epithelial Cells Exposed to IOERT. Anticancer Res 2015;35:3223-34. [PubMed]
- Bravatà V, Minafra L, Russo G, et al. High-dose Ionizing Radiation Regulates Gene Expression Changes in the MCF7 Breast Cancer Cell Line. Anticancer Res 2015;35:2577-91. [PubMed]
- Burnet NG, Barnett GC, Elliott RM, et al. RAPPER: the radiogenomics of radiation toxicity. Clin Oncol (R Coll Radiol) 2013;25:431-4. [Crossref] [PubMed]
- Guo Z, Shu Y, Zhou H, et al. Radiogenomics helps to achieve personalized therapy by evaluating patient responses to radiation treatment. Carcinogenesis 2015;36:307-17. [Crossref] [PubMed]
- Eke I, Makinde AY, Aryankalayil MJ, et al. Comprehensive molecular tumor profiling in radiation oncology: How it could be used for precision medicine. Cancer Lett 2016;382:118-26. [Crossref] [PubMed]
- Bonner JA, Harari PM, Giralt J, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med 2006;354:567-78. [Crossref] [PubMed]
- Bonner JA, Harari PM, Giralt J, et al. Radiotherapy plus cetuximab for locoregionally advanced head and neck cancer: 5-year survival data from a phase 3 randomised trial, and relation between cetuximab-induced rash and survival. Lancet Oncol 2010;11:21-8. [Crossref] [PubMed]
- Dewdney A, Cunningham D, Tabernero J, et al. Multicenter randomized phase Ii clinical trial comparing neoadjuvant oxaliplatin, capecitabine, and preoperative radiotherapy with or without cetuximab followed by total mesorectal excision in patients with high-risk rectal cancer (EXPERT-C). J Clin. Oncol 2012;30:1620-7. [Crossref] [PubMed]
- Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497-500. [Crossref] [PubMed]
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. [Crossref] [PubMed]
- Komaki R, Allen PK, Wei X, et al. Adding erlotinib to chemoradiation improves overall survival but not progression-free survival in stage iii non-small cell lung cancer. Int J Radiat Oncol Biol Phys 2015;92:317-24. [Crossref] [PubMed]
- Iyengar P, Kavanagh BD, Wardak Z, et al. Phase II trial of stereotactic body radiation therapy combined with erlotinib for patients with limited but progressive metastatic non-small-cell lung cancer. J Clin Oncol 2014;32:3824-30. [Crossref] [PubMed]
- Lilenbaum R, Samuels M, Wang X, et al. A phase II study of induction chemotherapy followed by thoracic radiotherapy and erlotinib in poor-risk stage III non-small-cell lung cancer: results of CALGB 30605 (Alliance)/RTOG 0972 (NRG). J Thorac Oncol 2015;10:143-7. [Crossref] [PubMed]
- Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 2011;364:2507-16. [Crossref] [PubMed]
- Satzger I, Degen A, Asper H, et al. Serious skin toxicity with the combination of BRAF inhibitors and radiotherapy. J Clin Oncol 2013;31:e220-e222. [Crossref] [PubMed]
- Amornwichet N, Oike T, Shibata A, et al. The EGFR mutation status affects the relative biological effectiveness of carbon-ion beams in non-small cell lung carcinoma cells. Sci Rep 2015;5:11305. [Crossref] [PubMed]
- Sologuren I, Rodríguez-Gallego C, Lara PC. Immune effects of high dose radiation treatment: implications of ionizing radiation on the development of bystander and abscopal effects. Transl Cancer Res 2014;3:18-31.
- Di Maggio FM, Minafra L, Forte GI, et al. Portrait of inflammatory response to ionizing radiation treatment. J Inflamm (Lond) 2015;12:14. [Crossref] [PubMed]
- Minafra L, Bravatà V. Cell and molecular response to IORT treatment. Transl Cancer Res 2014;3:32-47.
- Herskind C, Talbot CJ, Kerns SL, et al. Radiogenomics: A systems biology approach to understanding genetic risk factors for radiotherapy toxicity? Cancer Lett 2016;382:95-109. [Crossref] [PubMed]
- Unkel S, Belka C, Lauber K. On the analysis of clonogenic survival data: Statistical alternatives to the linear-quadratic model. Radiat Oncol 2016;11:11. [Crossref] [PubMed]
- Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 1994;367:645-8. [Crossref] [PubMed]
- Ailles LE, Weissman IL. Cancer stem cells in solid tumors. Curr Opin Biotechnol 2007;18:460-6. [Crossref] [PubMed]
- Sengupta A, Cancelas JA. Cancer stem cells: a stride towards cancer cure. J Cell Physiol 2010;225:7-14. [Crossref] [PubMed]
- Visvader JE. Cells of origin in cancer. Nature 2011;469:314-22. [Crossref] [PubMed]
- Dick JE. Looking a head in cancer stem cell research. Nat. Biotechnol 2009;27:44-6. [Crossref] [PubMed]
- Tomasetti C, Vogelstein B. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 2015;347:78-81. [Crossref] [PubMed]
- Odoux C, Fohrer H, Hoppo T, et al. A stochastic model for cancer stem cell origin in metastatic colon cancer. Cancer Res 2008;68:6932-41. [Crossref] [PubMed]
- Kaseb HO, Fohrer-Ting H, Lewis DW, et al. Identification, expansion and characterization of cancer cells with stem cell properties from head and neck squamous cell carcinomas. Exp Cell Res 2016;348:75-86. [Crossref] [PubMed]
- Pajonk F, Vlashi E, McBride WH. Radiation resistance of cancer stem cells: the 4 R's of radiobiology revisited. Stem Cells 2010;28:639-48. [Crossref] [PubMed]
- Sun L, Cabarcas SM and Farrar WL. Radioresistance and Cancer Stem Cells: Survival of the Fittest. J Carcinogene Mutagene 2011;S1:004.
- Skvortsova I, Debbageb P, Kumarc V, et al. Radiation resistance: cancer stem cells (CSCs) and their enigmatic pro-survival signaling. Semin Cancer Biol 2015;35:39-44. [Crossref] [PubMed]
- Gerweck LE, Wakimoto H. At the Crossroads of Cancer Stem Cells, Radiation Biology, and Radiation Oncology. Cancer Res 2016;76:994-8. [Crossref] [PubMed]
- Chang L, Graham P, Hao J, et al. Cancer stem cells and signaling pathways in radioresistance. Oncotarget 2016;7:11002-17. [PubMed]
- Bütof R, Dubrovska A, Baumann M. Clinical perspectives of cancer stem cell research in radiation oncology. Radiother Oncol 2013;108:388-96. [Crossref] [PubMed]
- Ropolo M, Daga A, Griffero F, et al. Comparative analysis of DNA repair in stem and nonstem glioma cell cultures. Mol Cancer Res 2009;7:383-92. [Crossref] [PubMed]
- Zhou W, Sun M, Li GH, et al. Activation of the phosphorylation of ATM contributes to radioresistance of glioma stem cells. Oncol Rep 2013;30:1793-801. [PubMed]
- Rainey MD, Charlton ME, Stanton RV, et al. Transient inhibition of ATM kinase is sufficient to enhance cellular sensitivity to ionizing radiation. Cancer Res 2008;68:7466-74. [Crossref] [PubMed]
- Tanei T, Morimoto K, Shimazu K, et al. Association of breast cancer stem cells identified by aldehyde dehydrogenase 1 expression with resistance to sequential Paclitaxel and epirubicin-based chemotherapy for breast cancers. Clin Cancer Res 2009;15:4234-41. [Crossref] [PubMed]
- Ahsan A, Hiniker SM, Davis MA, et al. Role of cell cycle in epidermal growth factor receptor inhibitor-mediated radiosensitization. Cancer Res 2009;69:5108-14. [Crossref] [PubMed]
- Hulleman E, Helin K. Molecular mechanisms in gliomagenesis. Adv Cancer Res 2005;94:1-27. [Crossref] [PubMed]
- Boyer MJ, Cheng T. The CDK inhibitors: potential targets for therapeutic stem cell manipulations. Gene Ther 2008;15:117-25. [Crossref] [PubMed]
- Phillips TM, McBride WH, Pajonk F. The response of CD24(-/low)/CD44+ breast cancer-initiating cells to radiation. J Natl Cancer Inst 2006;98:1777-85. [Crossref] [PubMed]
- Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006;444:756-60. [Crossref] [PubMed]
- Lim YC, Roberts TL, Day BW, et al. A role for homologous recombination and abnormal cell-cycle progression in radioresistance of glioma-initiating cells. Mol Cancer Ther 2012;11:1863-72. [Crossref] [PubMed]
- Zhang M, Behbod F, Atkinson RL, et al. Identification of tumor-initiating cells in a p53-null mouse model of breast cancer. Cancer Res 2008;68:4674-82. [Crossref] [PubMed]
- Weng AP, Aster JC. Multiple niches for Notch in cancer: context is everything. Curr Opin Genet Dev 2004;14:48-54. [Crossref] [PubMed]
- Takebe N, Miele L, Harris PJ, et al. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat Rev Clin Oncol 2015;12:445-464. [Crossref] [PubMed]
- Hittelman WN, Liao Y, Wang L, et al. Are cancer stem cells radioresistant? Future Oncol 2010;6:1563-76. [Crossref] [PubMed]
- Theys J, Yahyanejad S, Habets R, et al. High NOTCH activity induces radiation resistance in non small cell lung cancer. Radiother Oncol 2013;108:440-5. [Crossref] [PubMed]
- Wang J, Wakeman TP, Lathia JD, et al. Notch promotes radioresistance of glioma stem cells. Stem cells 2010;28:17-28. [PubMed]
- Gupta R, Vyas P, Enver T. Molecular targeting of cancer stem cells. Cell Stem Cell 2009;5:125-6. [Crossref] [PubMed]
- Hanna A, Shevde LA. Hedgehog signaling: modulation of cancer properties and tumor mircroenvironment. Mol Cancer 2016;15:24. [Crossref] [PubMed]
- Matsui WH. Cancer stem cell signaling pathways. Medicine (Baltimore) 2016;95:S8-S19. [Crossref] [PubMed]
- Clement V, Sanchez P, de Tribolet N, et al. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol 2007;17:165-72. [Crossref] [PubMed]
- Sims-Mourtada J, Izzo JG, Apisarnthanarax S, et al. Hedgehog: an attribute to tumor regrowth after chemoradiotherapy and a target to improve radiation response. Clin Cancer Res 2006;12:6565-72. [Crossref] [PubMed]
- Gan GN, Eagles J, Keysar SB, et al. Hedgehog signaling drives radioresistance and stroma-driven tumor repopulation in head and neck squamous cancers. Cancer Res 2014;74:7024-36. [Crossref] [PubMed]
- Li X, Chen F, Zhu Q, et al. Gli-1/PI3K/AKT/NF-kB pathway mediates resistance to radiation and is a target for reversion of responses in refractory acute myeloid leukemia cells. Oncotarget 2016;7:33004-15. [PubMed]
- Takebe N, Ivy SP. Controversies in cancer stem cells: targeting embryonic signaling pathways. Clin Cancer Res 2010;16:3106-12. [Crossref] [PubMed]
- Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell 2012;149:1192-205. [Crossref] [PubMed]
- Woodward WA, Chen MS, Behbod F, et al. WNT/beta-catenin mediates radiation resistance of mouse mammary progenitor cells. Proc Natl Acad Sci U S A 2007;104:618-23. [Crossref] [PubMed]
- Chang HW, Nam HY, Kim HJ, et al. Effect of β-catenin silencing in overcoming radioresistance of head and neck cancer cells by antagonizing the effects of AMPK on Ku70/Ku80. Head Neck 2016;38:E1909-17. [Crossref] [PubMed]
- Ischenko I, Seeliger H, Schaffer M, et al. Cancer stem cells: how can we target them. Curr Med Chem 2008;15:3171-84. [Crossref] [PubMed]
- Bayer C, Shi K, Astner ST, et al. Acute versus chronic hypoxia: why a simplified classification is simply not enough. Int J Radiat Oncol Biol Phys 2011;80:965-8. [Crossref] [PubMed]
- Span PN, Bussink J. Biology of hypoxia. Semin Nucl Med 2015;45:101-9. [Crossref] [PubMed]
- Liu C, Lin Q, Yun Z. Cellular and Molecular Mechanisms Underlying Oxygen-Dependent Radiosensitivity. Radiat Res 2015;183:487-96. [Crossref] [PubMed]
- Horsman MR, Overgaard J. The impact of hypoxia and its modification of the outcome of radiotherapy. J Radiat Res 2016;57:i90-i98. [Crossref] [PubMed]
- Brown JM, Wilson WR. Exploiting tumour hypoxia in cancer treatment. Nat Rev Cancer 2004;4:437-47. [Crossref] [PubMed]
- Hubbi ME, Semenza GL. Regulation of cell proliferation by hypoxia-inducible factors. Am J Physiol Cell Physiol 2015;309:C775-82. [Crossref] [PubMed]
- Wigerup C, Pahlman S, Bexell D. Therapeutic targeting of hypoxia and hypoxia-inducible factors in cancer. Pharmacol Ther 2016;164:152-69. [Crossref] [PubMed]
- Semenza GL. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J Clin Invest 2013;123:3664-71. [Crossref] [PubMed]
- Harada H, Inoue M, Itasaka S, et al. Cancer cells that survive radiation therapy acquire HIF-1 activity and translocate towards tumour blood vessels. Nat Commun 2012;3:783. [Crossref] [PubMed]
- Harris BH, Barberis A, West CM, et al. Gene Expression Signatures as Biomarkers of Tumour Hypoxia. Clin Oncol (R Coll Radiol) 2015;27:547-60. [Crossref] [PubMed]
- Harada H, Itasaka S, Zhu Y, et al. Treatment regimen determines whether an HIF-1 inhibitor enhances or inhibits the effect of radiation therapy. Br J Cancer 2009;100:747-57. [Crossref] [PubMed]
- Meijer TW, Kaanders JH, Span PN, et al. Targeting hypoxia, HIF-1, and tumor glucose metabolism to improve radiotherapy efficacy. Clin Cancer Res 2012;18:5585-94. [Crossref] [PubMed]
- Wilson WR, Hay MP. Targeting hypoxia in cancer therapy. Nat Rev Cancer 2011;11:393-410. [Crossref] [PubMed]
- Ding S, Li C, Cheng N, et al. Redox Regulation in Cancer Stem Cells. Oxid Med Cell Longev 2015;2015:750798 [Crossref] [PubMed]
- Borovski T, De Sousa EM, Vermeulen L, et al. Cancer stem cell niche: the place to be. Cancer Res 2011;71:634-9. [Crossref] [PubMed]
- Ghotra VP, Puigvert JC, Danen EH. The cancer stem cell microenvironment and anti-cancer therapy. Int J Radiat Biol 2009;85:955-62. [Crossref] [PubMed]
- Rödel F, Frey B, Leitmann W, et al. Survivin antisense oligonucleotides effectively radiosensitize colorectal cancer cells in both tissue culture and murine xenograft models. Int J Radiat Oncol Biol Phys 2008;71:247-55. [Crossref] [PubMed]
- Lomonaco SL, Finniss S, Xiang C, et al. The induction of autophagy by gamma-radiation contributes to the radioresistance of glioma stem cells. Int J Cancer 2009;125:717-22. [Crossref] [PubMed]
- Blazek ER, Foutch JL, Maki G. Daoy medulloblastoma cells that express CD133 are radioresistant relative to CD133-cells, and the CD133+ sector is enlarged by hypoxia. Int J Radiat Oncol Biol Phys 2007;67:1-5. [Crossref] [PubMed]
- Hovinga KE, Shimizu F, Wang R, et al. Inhibition of notch signaling in glioblastoma targets cancer stem cells via an endothelial cell intermediate. Stem Cells 2010;28:1019-29. [Crossref] [PubMed]
- Cenciarelli C, Marei HE, Zonfrillo M, et al. PDGF receptor alpha inhibition induces apoptosis in glioblastoma cancer stem cells refractory to anti-Notch and anti-EGFR treatment. Mol Cancer 2014;13:247. [Crossref] [PubMed]
- Fan X, Khaki L, Zhu TS, et al. NOTCH pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts. Stem Cells 2010;28:5-16. [PubMed]
- Soeda A, Park M, Lee D, et al. Hypoxia promotes expansion of the CD133-positive glioma stem cells through activation of HIF-1 alpha. Oncogene 2009;28:3949-59. [Crossref] [PubMed]
- Lee G, Hall RR 3rd, Ahmed AU. Cancer Stem Cells: Cellular Plasticity, Niche, and its Clinical Relevance. J Stem Cell Res Ther 2016;6:363 [Crossref] [PubMed]
- Potten CS. Cell cycles in cell hierarchies. Int J Radiat Biol Relat Stud Phys Chem Med 1986;49:257-78. [Crossref] [PubMed]
- Weinberg RA. The Biology of Cancer, second ed. Garland Science Publishing, New York, Oxford. 2013:960.
- Pajonk F, Vlashi E. Characterization of the stem cell niche and its importance in radiobiological response. Semin Radiat Oncol 2013;23:237-41. [Crossref] [PubMed]
- Dörr W. Three A’s of repopulation during fractionated irradiation of squamous epithelia: asymmetry loss, acceleration of stem-cell divisions and abortive divisions Int J Radiat Biol 1997;72:635-43. [Crossref] [PubMed]
- Stewart FA, Dorr W. Milestones in normal tissue radiation biology over the past 50 years: from clonogenic cell survival to cytokine networks and back to stem cell recovery. Int J Radiat Biol 2009;85:574-86. [Crossref] [PubMed]
- West CM, Barnett GC. Genetics and genomics of radiotherapy toxicity: towards prediction. Genome Med 2011;3:52. [Crossref] [PubMed]
- Bayreuther K, Francz PI, Rodemann HP. Fibroblasts in normal and pathological terminal differentiation, aging, apoptosis and transformation. Arch Gerontol Geriatr 1992;15:47-74. [Crossref] [PubMed]
- Rodemann HP, Peterson HP, Schwenke K, et al. Terminal differentiation of human fibroblasts is induced by radiation. Scanning Microsc 1991;5:1135-42. [PubMed]
- Delanian S, Martin M, Bravard A, et al. Abnormal phenotype of cultured fibroblasts in human skin with chronic radiotherapy damage. Radiother Oncol 1998;47:255-61. [Crossref] [PubMed]
- Safwat A, Bentzen SM, Turesson I, et al. Deterministic rather than stochastic factors explain most of the variation in the expression of skin telangiectasia after radiotherapy. Int J Radiat Oncol Biol Phys 2002;52:198-204. [Crossref] [PubMed]
- Tucker SL, Turesson I, Thames HD. Evidence for individual differences in the radiosensitivity of human skin. Eur J Cancer 1992;28A:1783-91. [Crossref] [PubMed]
- Giotopoulos G, Symonds RP, Foweraker K, et al. The late radiotherapy normal tissue injury phenotypes of telangiectasia, fibrosis and atrophy in breast cancer patients have distinct genotype-dependent causes. Br J Cancer 2007;96:1001-7. [Crossref] [PubMed]
- Barnett GC, Coles CE, Burnet NG, et al. No association between SNPs regulating TGF-beta1 secretion and late radiotherapy toxicity to the breast: results from the RAPPER study. Radiother Oncol 2010;97:9-14. [Crossref] [PubMed]
- Grantzau T, Overgaard J. Risk of second non-breast cancer after radiotherapy for breast cancer: a systematic review and meta-analysis of 762,468 patients. Radiother Oncol 2015;114:56-65. [Crossref] [PubMed]
- Aypar U, Morgan WF, Baulch JE. Radiation-induced genomic instability: are epigenetic mechanisms the missing link? Int J Radiat Biol 2011;87:179-91. [Crossref] [PubMed]
- Morgan WF. Radiation-induced genomic instability. Health Phys 2011;100:280-1. [Crossref] [PubMed]
- Pollard JM, Gatti RA. Clinical radiation sensitivity with DNA repair disorders: an overview. Int J Radiat Oncol Biol Phys 2009;74:1323-31. [Crossref] [PubMed]
- Andreassen CN. Can risk of radiotherapy-induced normal tissue complications be predicted from genetic profiles? Acta Oncol 2005;44:801-15. [Crossref] [PubMed]
- Andreassen CN, Schack LM, Laursen LV, et al. Radiogenomics-current status, challenges and future directions. Cancer Lett. 2016;382:127-36. [Crossref] [PubMed]
- Talbot CJ, Tanteles GA, Barnett GC, et al. A replicated association between polymorphisms near TNF-alpha and risk for adverse reactions to radiotherapy. Br J Cancer 2012;107:748-53. [Crossref] [PubMed]
- Edvardsen H, Landmark-Hoyvik H, Reinertsen KV, et al. SNP in TXNRD2 associated with radiation induced fibrosis: a study of genetic variation in reactive oxygen species metabolism and signaling. Int J Radiat Oncol Biol Phys 2013;86:791-9. [Crossref] [PubMed]
- Seibold P, Behrens S, Schmezer P, et al. XRCC1 polymorphism associated with late toxicity after radiation therapy in breast cancer patients. Int J Radiat Oncol Biol Phys 2015;92:1084-92. [Crossref] [PubMed]
- Rosenstein BS, West CM, Bentzen SM, et al. Radiogenomics: radiobiology enters the era of big data and team science. Int J Radiat Oncol Biol Phys 2014;89:709-13. [Crossref] [PubMed]
- West C, Rosenstein BS, Alsner J, et al. Establishment of a radiogenomics consortium. Int J Radiat Oncol Biol Phys 2010;76:1295-6. [Crossref] [PubMed]
- Andreassen CN, Barnett GC, Kerns SL, et al. OC-0143: analysis of 5434 patients shows a link between the ATM codon 1853 SNP and the risk of radiation-induced toxicity. Radiother Oncol 2014;111:S55-S56. [Crossref]
- Minafra L, Bravatà V, Saporito M, et al. Genetic, clinical and radiographic signs in knee osteoarthritis susceptibility. Arthritis Res Ther 2014;16:R91. [Crossref] [PubMed]
- Bravatà V, Minafra L, Forte GI, et al. DVWA gene polymorphisms and osteoarthritis. BMC Res Notes 2015;8:30. [Crossref] [PubMed]
- Barnett GC, Thompson D, Fachal L, et al. A genome wide association study (GWAS) providing evidence of an association between common genetic variants and late radiotherapy toxicity. Radiother Oncol 2014;111:178-85. [Crossref] [PubMed]
- Fachal L, Gómez-Caamaño A, Barnett GC, et al. A three-stage genome-wide association study identifies a susceptibility locus for late radiotherapy toxicity at 2q24.1. Nat Genet 2014;46:891-4. [Crossref] [PubMed]
- Ahmed M, Dorling L, Kerns S, et al. Common genetic variation associated with increased susceptibility to prostate cancer does not increase risk of radiotherapy toxicity. Br J Cancer 2016;114:1165-74. [Crossref] [PubMed]
- Kerns SL, Ostrer H, Stock RG, et al. Genome-wide association study to identify single nucleotide polymorphisms (SNPs) associated with the development of erectile dysfunction in African-American men after radiotherapy for prostate cancer. Int J Radiat Oncol Biol Phys 2010;78:1292-300. [Crossref] [PubMed]
- Kerns SL, Stock R, Stone N, et al. A 2-stage genome-wide association study to identify single nucleotide polymorphisms associated with development of erectile dysfunction following radiation therapy for prostate cancer. Int J Radiat Oncol Biol Phys 2013;85:e21-8. [Crossref] [PubMed]
- Kerns SL, Stock RG, Stone NN, et al. Genome-wide association study identifies a region on chromosome 11q14.3 associated with late rectal bleeding following radiation therapy for prostate cancer. Radiother Oncol 2013;107:372-6. [Crossref] [PubMed]
- Kerns SL, Stone NN, Stock RG, et al. A 2-stage genome-wide association study to identify single nucleotide polymorphisms associated with development of urinary symptoms after radiotherapy for prostate cancer. J Urol 2013;190:102-8. [Crossref] [PubMed]
- Alsbeih GA, Al-Harbi NM, El-Sebaie MM, et al. Involvement of mitochondrial DNA sequence variations and respiratory activity in late complications following radiotherapy. Clin Cancer Res 2009;15:7352-60. [Crossref] [PubMed]
- Roberson JD, Burnett OL. Radiogenomics: towards a personalized radiation oncology. Curr Opin Pediatr 2016;28:713-7. [Crossref] [PubMed]
- Edwards SL, Beesley J, French JD, et al. Beyond GWASs: illuminating the dark road from association to function. Am J Hum Genet 2013;93:779-97. [Crossref] [PubMed]
- Albert FW, Kruglyak L. The role of regulatory variation in complex traits and disease. Nat Rev Genet 2015;16:197-212. [Crossref] [PubMed]
- Westra HJ, Franke L. From genome to function by studying eQTLs. Biochim Biophys Acta 2014;1842:1896-902. [Crossref] [PubMed]
- Smirnov DA, Morley M, Shin E, et al. Genetic analysis of radiation-induced changes in human gene expression. Nature 2009;459:587-91. [Crossref] [PubMed]
- Niu N, Qin Y, Fridley BL, et al. Radiation pharmacogenomics: a genome-wide association approach to identify radiation response biomarkers using human lymphoblastoid cell lines. Genome Res 2010;20:1482-92. [Crossref] [PubMed]
- Rieger KE, Hong WJ, Tusher VG, et al. Toxicity from radiation therapy associated with abnormal transcriptional responses to DNA damage. Proc Natl Acad Sci U S A 2004;101:6635-40. [Crossref] [PubMed]
- Svensson JP, Stalpers LJ, Esveldt-van Lange RE, et al. Analysis of gene expression using gene sets discriminates cancer patients with and without late radiation toxicity. PLoS Med 2006;3:e422 [Crossref] [PubMed]
- Mayer C, Popanda O, Greve B, et al. A radiation-induced gene expression signature as a tool to predict acute radiotherapy-induced adverse side effects. Cancer Lett 2011;302:20-8. [Crossref] [PubMed]
- Greve B, Bolling T, Amler S, et al. Evaluation of different biomarkers to predict individual radiosensitivity in an interlaboratory comparison-lessons for future studies. PLoS One 2012;7:e47185 [Crossref] [PubMed]
- Finnon P, Kabacik S, MacKay A, et al. Correlation of in vitro lymphocyte radiosensitivity and gene expression with late normal tissue reactions following curative radiotherapy for breast cancer. Radiother Oncol 2012;105:329-36. [Crossref] [PubMed]
- van Oorschot B, Hovingh SE, Moerland PD, et al. Reduced activity of double-strand break repair genes in prostate cancer patients with late normal tissue radiation toxicity. Int J Radiat Oncol Biol Phys 2014;88:664-70. [Crossref] [PubMed]
- Koerdt S, Tanner N, Rommel N, et al. NOS1-, NOS3-, PIK3CA-, and MAPK-pathways in skin following radiation therapy. Biomark Res 2017;5:3. [Crossref] [PubMed]
- Landmark-Høyvik H, Dumeaux V, Reinertsen KV, et al. Blood gene expression profiling of breast cancer survivors experiencing fibrosis. Int J Radiat Oncol Biol Phys 2011;79:875-83. [Crossref] [PubMed]
- Ghosh AK, Vaughan DE. PAI-1 in tissue fibrosis. J. Cell. Physiol 2012;227:493-507. [Crossref] [PubMed]
- Rødningen OK, Overgaard J, Alsner J, et al. Microarray analysis of the transcriptional response to single or multiple doses of ionizing radiation in human subcutaneous fibroblasts. Radiother Oncol 2005;77:231-40. [Crossref] [PubMed]
- Alsner J, Rodningen OK, Overgaard J. Differential gene expression before and after ionizing radiation of subcutaneous fibroblasts identifies breast cancer patients resistant to radiation-induced fibrosis. Radiother Oncol 2007;83:261-6. [Crossref] [PubMed]
- Rødningen OK, Borresen-Dale AL, Alsner J, et al. Radiation induced gene expression in human subcutaneous fibroblasts is predictive of radiation-induced fibrosis. Radiother Oncol 2008;86:314-20. [Crossref] [PubMed]
- Andreassen CN, Overgaard J, Alsner J. Independent prospective validation of a predictive test for risk of radiation induced fibrosis based on the gene expression pattern in fibroblasts irradiated in vitro. Radiother Oncol 2013;108:469-72. [Crossref] [PubMed]
- Stenmark MH, Cai XW, Shedden K, et al. Combining physical and biologic parameters to predict radiation-induced lung toxicity in patients with non-small-cell lung cancer treated with definitive radiation therapy. Int J Radiat Oncol Biol Phys 2012;84:e217-e222. [Crossref] [PubMed]
- Anscher MS, Kong FM, Andrew K, et al. Plasma transforming growth factor beta1 as a predictor of radiation pneumonitis. Int J Radiat Oncol Biol Phys 1998;41:1029-35. [Crossref] [PubMed]
- Novakova-Jiresova A, Van Gameren MM, Coppes RP, et al. Transforming growth factor-beta plasma dynamics and post-irradiation lung injury in lung cancer patients. Radiother Oncol 2004;71:183-9. [Crossref] [PubMed]
- Rübe CE, Palm J, Erren M, et al. Cytokine plasma levels: reliable predictors for radiation pneumonitis? PLoS One 2008;3:e2898 [Crossref] [PubMed]
- Zhao L, Sheldon K, Chen M, et al. The predictive role of plasma TGF-beta1 during radiation therapy for radiation induced lung toxicity deserves further study in patients with non-small cell lung cancer. Lung Cancer 2008;59:232-9. [Crossref] [PubMed]
- De Jaeger K, Seppenwoolde Y, Kampinga HH, et al. Significance of plasma transforming growth factor-beta levels in radiotherapy for non-small-cell lung cancer. Int J Radiat Oncol Biol Phys 2004;58:1378-87. [Crossref] [PubMed]
- Dickson J, Magee B, Stewart A, et al. Relationship between residual radiation-induced DNA double-strand breaks in cultured fibroblasts and late radiation reactions: a comparison of training and validation cohorts of breast cancer patients. Radiother Oncol 2002;62:321-6. [Crossref] [PubMed]
- Oh JH, Craft JM, Townsend R, et al. A bioinformatics approach for biomarker identification in radiation-induced lung inflammation from limited proteomics data. J Proteome Res 2011;10:1406-15. [Crossref] [PubMed]
- Cai XW, Shedden KA, Yuan SH, et al. Baseline plasma proteomic analysis to identify biomarkers that predict radiation induced lung toxicity in patients receiving radiation for non-small cell lung cancer. J Thorac Oncol 2011;6:1073-8. [Crossref] [PubMed]
- Pathak R, Cheema AK, Boca SM, et al. Modulation of Radiation Response by the Tetrahydrobiopterin Pathway. Antioxidants (Basel) 2015;4:68-81. [Crossref] [PubMed]
- Cook JA, Chandramouli GV, Anver MR, et al. Mass Spectrometry-Based Metabolomics Identifies Longitudinal Urinary Metabolite Profiles Predictive of Radiation-Induced Cancer. Cancer Res 2016;76:1569-77. [Crossref] [PubMed]
- Menon SS, Uppal M, Randhawa S, et al. Radiation Metabolomics: Current Status and Future Directions. Front Oncol 2016;6:20. [Crossref] [PubMed]
- Xu T, Liao Z, O'Reilly MS, et al. Serum inflammatory miRNAs predict radiation esophagitis in patients receiving definitive radiochemotherapy for non-small cell lung cancer. Radiother Oncol 2014;113:379-84. [Crossref] [PubMed]
- Weigel C, Veldwijk MR, Oakes CC, et al. Epigenetic regulation of diacylglycerol kinase alpha promotes radiation-induced fibrosis. Nat Commun 2016;7:10893. [Crossref] [PubMed]
- Weigel C, Schmezer P, Plass C, et al. Epigenetics in radiation-induced fibrosis. Oncogene 2015;34:2145-55. [Crossref] [PubMed]
- Paulíková S, Chmelařová M, Petera J, et al. Hypermethylation of RAD51L3 and XRCC2 genes to predict late toxicity in chemoradiotherapy-treated cervical cancer patients. Folia Biol (Praha) 2013;59:240-5. [PubMed]
- Simone BA, Ly D, Savage JE, et al. MicroRNA alterations driving acute and late stages of radiation-induced fibrosis in a murine skin model. Int J Radiat Oncol Biol Phys 2014;90:44-52. [Crossref] [PubMed]
- Someya M, Yamamoto H, Nojima M, et al. Relation between Ku80 and microRNA-99a expression and late rectal bleeding after radiotherapy for prostate cancer. Radiother Oncol 2015;115:235-9. [Crossref] [PubMed]