
Hyperpolarization-activated cyclic nucleotide-gated gene signatures and poor clinical outcome of cancer patient
Introduction
Cancer is the leading cause of death all over the globe in recent decades. According to WHO, there were 8.2 million people who died from cancer in the year 2012, and in the next two decades this figure will grow to around 22 million (1). Until now, only 30% of cancers could be prevented (1). Commonly, cancer patients would undergo surgery synergistically with chemotherapy and/or radiotherapy, which is painful, and has high mortality rate. According to the U.S. National Cancer Institute, around 200 cancer drugs are commercialized in the market. Nevertheless, researchers in cancer field are still looking to develop new anti-cancer drugs with more specificity and high efficiency (2).
Hyperpolarization-activated cyclic nucleotide-gated (HCN) channel is one of hundreds of intra-membrane ion channels involved in ion transport. HCN channels are encoded by four genes, namely HCN1, HCN2, HCN3 and HCN4 (3). These four genes are predominantly localized and expressed in the heart and the central nervous system (3,4). HCN channels are activated by hyperpolarization, and which permit Na+ and K+ to flow inward to the cell (5). HCN channels’ main physiological functions are in the heart (6) and the nervous system (4). HCN genes were found to play a role in arrhythmogenic disease and neurological disease (7). The pharmacological properties of these ion channels in cancer are relatively unknown.
Oncomine is a web-based database, which contains more than 700 independent datasets with an estimated 90,000 microarray trials (8,9). The use of Oncomine in several publications confirmed it is a reliable source of clinical datasets (10-16). Oncomine standardizes and organizes the datasets of public cancer microarray data into different cancer types and subtypes (8,9).
NextBio Research database (Illumina INC.) is a web-based platform containing microarray data of more than 20,000 published studies. This online database was introduced by Giovanni Coppola in his book in 2013 (17) and has been used in previous studies (18,19).
In this study, data mining of Oncomine and NextBio Research database was performed to conduct a meta-analysis of HCN gene expression across multiple types and subtypes of cancer. In addition, analysis of survival rate of cancer patients and HCN gene expression was conducted to investigate how these expressions affect the overall survival of cancer patients in the 3 and 5 years’ period.
Methods
Data mining
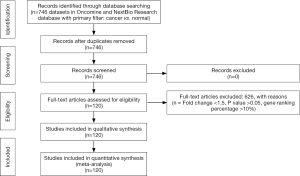
A meta-analysis was performed to analyze the mRNA expression level of HCN gene family in clinical cancer specimens following PRISMA guidelines (20,21) (Figure 1, Tables S1-S9).
HCN gene (HCN1-4) expression within 17 cancer types was investigated. The mRNA expression of HCN genes in cancerous tissues was grouped by origin of tissue and then compared to normal tissue. Oncomine (www.oncomine.org) and NextBio Research database (https://www.nextbio.com) were used to analyze the mRNA expression of HCN gene family in clinical cancer tissues (22).
Database search strategy
In this study, the cancer vs. normal filter was chosen, which only displayed datasets examining HCN gene mRNA expression in the same origin of tissue. In order to be included in the study, all the data from Oncomine and NextBio research database must satisfy the following threshold: P<0.05, a fold change >1.5 and a gene rank percentile <10% (only applicable to data from Oncomine) (9) (Figure 1). Statistical analyses were conducted with Oncomine and NextBio Research default algorithms such as P values, two-tailed Student’s t-test, and multiple testing corrections. In total, there were 120 studies with 8,471 samples included in this study. All the searches were performed from December 2015 to December 2016.
Survival analysis
The correlation between HCN gene family and overall survival rate was analyzed using Kaplan-Meier plotter (http://kmplot.com/) (23) and PROGgeneV2 (24). Two groups of patients were used for the comparison on survival rates with high and low expression levels of HCN1, HCN2, HCN3 and HCN4 gene.
All the searches were performed from December 2015 to December 2016.
Results
Expression of HCN1 in multiple types and subtypes of cancer
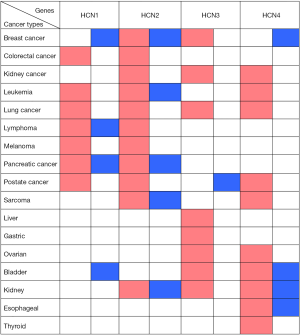
In general, HCN1 gene over-expressed in diverse types of cancer such as colorectal cancer, leukemia, lung cancer, melanoma, and prostate cancer whereas the mRNA expression of HCN1 was under-expressed in breast cancer and bladder cancer. In addition, HCN1 gene also over and under expressed in both lymphoma and pancreatic cancer (Figure 2).
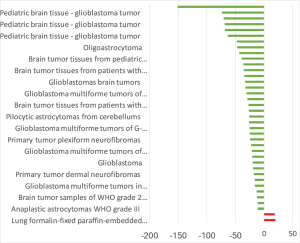
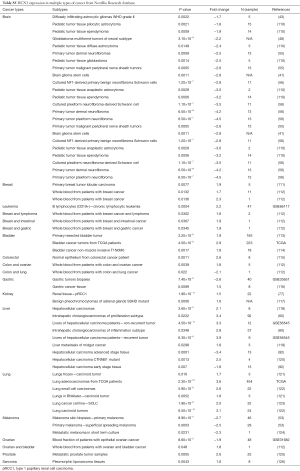
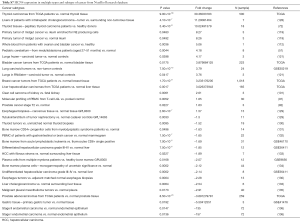
The highest expression fold change of HCN1 in cancer and normal matched type tissue was displayed in Figure 3. HCN1 expression in brain cancer particular in glioblastoma, glioma was extremely low with the lowest fold change of −151-fold relative to normal brain tissue. However, HCN1 expression was up-regulated in hepatocellular carcinoma and lung cancer with the fold change of 18.8 and 19.9-fold respectively.
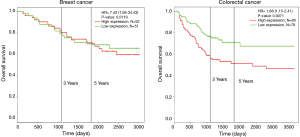
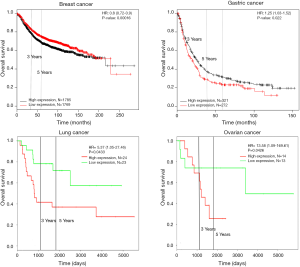
Survival analysis of HCN1 expression using Kaplan-Meier plotter and PROGgeneV2 showed that HCN1 had significant correlation with mortality in breast cancer [hazard ratio (HR) =7.42, P=0.0019] and colorectal cancer (HR =1.66, P=0.0071) (Figure 4).
Expression of HCN2 in multiple types and subtypes of cancer
In our study, we found that HCN2 gene overexpressed in colorectal cancer, kidney cancer, lung cancer, lymphoma, melanoma, and prostate cancer whereas HCN2 expression level showed both up and down regulation in breast cancer, leukemia, pancreatic cancer, sarcoma, and kidney cancer (Figure 2).
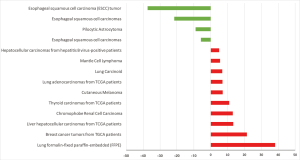
HCN2 fold change was calculated and displayed in Figure 5. HCN2 expression was down-regulated in esophageal squamous cell carcinoma with fold change of −37.3 and −21.6-fold relative to normal matched type tissue. In contrast, HCN2 was up-regulated in lung cancer, breast cancer, liver cancer, and thyroid cancer with 38.1, 21.4, 13.3, 10.9-fold respectively higher than normal control tissue.
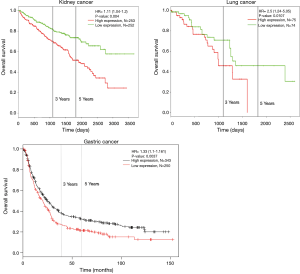
To further investigate the expression of HCN2 and overall survival rate of cancer patients, we used Kaplan-Meier plotter analysis and PROGgeneV2 and found that the lower survival rates of lung cancer (HR =2.5, P=0.0107), kidney cancer (HR =1.1, P=0.004) and gastric cancer (HR =1.33, P=0.0037) had significant correlation with the expression of HCN2 (Figure 6). The current data suggested that overexpression of HCN2 may be involved in the particular process of lung cancer. This observation may make HCN2 a potential biomarker for esophageal squamous cell carcinoma, lung cancer, kidney cancer and gastric cancer, breast cancer, liver cancer, and thyroid cancer diagnosis and prognosis.
Expression of HCN3 in multiple types and subtypes of cancer
Our data showed that HCN3 gene over-expressed in breast cancer, kidney cancer, lung cancer, liver cancer, gastric cancer, ovarian cancer, bladder cancer, kidney cancer whereas HCN3 was under-expressed in prostate cancer (Figure 2).
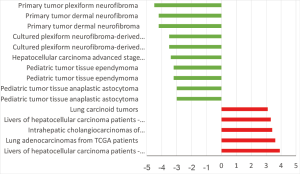
HCN3 was under expressed in many subtypes of brain cancer such as primary tumor dermal neurofibroma, cultured plexiform neurofibroma-derived Schwann cell, pediatic tumor tissue ependymoma, pediatic tumor tissue anaplastic astrocytoma with fold change of −4.5, −3.5, −3.2, −3-fold respectively compared to normal matched type tissue (Figure 7). In contrast, HCN3 over-expressed in liver and lung cancer tissue with fold change of 3.9 and 3.6 respectively, relatively to normal matched type sample (Figure 7).
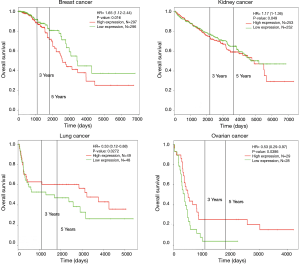
In addition, Kaplan-Meier plotter and PROGgeneV2 analysis showed overexpression of HCN3 in breast cancer was significantly correlated with lower survival rates and poor prognosis value of breast cancer (HR =1.65, P=0.0016), kidney cancer (HR =1.17, P=0.0049) but higher survival rate and good prognosis value in lung cancer (HR =0.33, P=0.0272) and ovarian cancer (HR =0.53, P=0.0386) patients (Figure 8). This result may indicate HCN3 as a potential biomarker for diagnosis and prognosis of brain cancer, breast cancer, kidney cancer, lung cancer and ovarian cancer.
Expression of HCN4 in multiple types and subtypes of cancer
HCN4 gene was found over-expression in kidney cancer, leukemia, lung cancer, sarcoma, ovarian cancer, and thyroid cancer whereas it under expressed in breast cancer. In addition, both over and under expression of HCN4 were found in bladder cancer, kidney cancer, and esophageal cancer (Figure 2).
HCN4 was under-expressed in stage I and II endometrial carcinoma with the fold change extremely low (−157 and −135-fold). In contrast, HCN4 was over expressed in Thyroid carcinomas, Thyroid tissues-papillary thyroid carcinoma, liver cancer, and prostate cancer with fold change of 30.4, 10, 11.2, and 7.8-fold respectively (Figure 9).
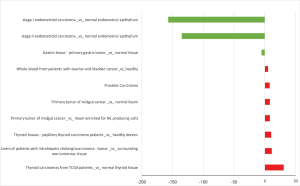
Kaplan-Meier plotter analysis showed that upregulation of HCN4 was highly correlated with the lower survival rates of patients with gastric cancer (HR =1.25, P=0.022), lung cancer (HR =5.37, P=0.0433) and ovarian cancer (HR =13.58, P=0.0426) but higher survival rate in patient with breast cancer (HR =0.8, P=0.00016) (Figure 10). From these results, HCN4 can be considered as the potential marker in breast cancer, gastric cancer, lung cancer and ovarian cancer diagnosis, thyroid carcinomas.
Discussion
In this study, we showed that HCN family members (HCN1, HCN2, HCN3, HCN4) overexpressed in numerous cancerous tissue relative to normal matched tissue. The increased expression of these four genes in multiple types and subtypes of cancer was also significantly correlated with low and high survival rates of cancer patients. This correlation suggests that HCN genes might play a key role in cancer particularly in brain cancer, lung cancer, liver cancer, esophageal cancer, thyroid cancer, ovarian cancer which had notably high fold change compared to normal matched type tissue. However, further study is required to confirm the mechanism of how HCN genes play a role in cancer.
In neuropathic pain, lacking of HCN1 gene expression by genetic deletion showed mitigation in neuronal damage (25). Another research showed that HCN1 deficiency caused epilepsy, ataxia and learning compromise (7). In a recent study, HCN1 showed under-expression with fold change of 0.65 in breast cancer cells after Maitake D-Fraction treatment (26). Single nucleotide polymorphism of HCN1 was found association with shorter survival of breast cancer patient (27). Moreover, inhibiting of HCN channel functions in embryonic stem cells by ZD7288, a HCN channels blocker, and cesium revealed that cell proliferation was decreased under the effects of these two drugs (28). HCN3 gene was also implied as the potential target for tumor suppression (28). In our findings, HCN3 also showed overexpression in multiple types of cancers such as breast cancer, and liver cancer. Therefore, we speculate that HCN3 is likely to be a candidate to study cancer cell proliferation and cancer cell cycle.
The HCN gene is commonly located in ventricular myocytes and neuron cells. Previous studies on the roles of the HCN channels were primarily focused on neurological diseases such as epilepsies and neuropathic pain disorders and cardiac related diseases (7,29,30). HCN channel functions are largely unknown in cancer. HCN channels (HCN1-4) have been known to allow the flow of Na+ and K+ ions (1:4 ration) inward and outward of the cell, which creates a hyperpolarization activated current named Ih. This current was showed to participate in regulating the heart rate and the firing of neurons. Moreover, HCN channels also play a role in the determination of resting membrane potential, dendritic integration, synaptic transmission and learning (7). HCN2 roles in inflammatory and neuropathic pain have been uncovered recently (31). Intriguingly, apart from the permeability of Na+ and K+ inward cell, HCN2 and HCN4 also allow Ca2+ ion into the cell (32). This happened due to the dephosphorylation of Thr549 within the regulatory region of HCN2; and calcium ion influx causes cell apoptosis due to cytotoxicity (33). cAMP was acknowledged to modulate HCN2 in gating activity (34,35). Moreover, in non-small cell lung carcinomas, HCN2 has also been triggered by PKC inhibitors such as staurosporine (STS) or PKC412 and under expression of HCN2 can prevent cell apoptosis (36). As a consequence, if HCN2 is mutated or overexpressed in cancer cells, it can lead to cancer cell not going through apoptosis. Thus, HCN2 expression is crucial and likely to be a potential target for cancer treatment via inhibiting of HCN2 expression.
The current study is the pioneer meta-analysis research about HCN gene expression in multiple types and subtypes of cancer. HCN1-4 could have potency as biomarker for cancer disease diagnosis and prognosis. Further study on HCN genes and specific types of cancer as suggested in the present study may help to reveal the underlying molecular mechanism of these genes in cancer.
Full table

Full table
Full table

Full table
Full table

Full table
Full table
Full table

Full table
Acknowledgments
Funding: This project is supported by National Science Council of Yuan (NSC 104-2320-B-034-003; NSC 105-2320-B-034-001 to YC Lin).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.07.22). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Zugazagoitia J, Guedes C, Ponce S, et al. Current Challenges in Cancer Treatment. Clin Ther 2016;38:1551-66. [Crossref] [PubMed]
- Kaupp UB, Seifert R. Molecular diversity of pacemaker ion channels. Annu Rev Physiol 2001;63:235-57. [Crossref] [PubMed]
- Notomi T, Shigemoto R. Immunohistochemical localization of Ih channel subunits, HCN1-4, in the rat brain. J Comp Neurol 2004;471:241-76. [Crossref] [PubMed]
- Bender RA, Baram TZ. Hyperpolarization activated cyclic-nucleotide gated (HCN) channels in developing neuronal networks. Prog Neurobiol 2008;86:129-40. [Crossref] [PubMed]
- Larsson HP. How is the heart rate regulated in the sinoatrial node? Another piece to the puzzle. J Gen Physiol 2010;136:237-41. [Crossref] [PubMed]
- Postea O, Biel M. Exploring HCN channels as novel drug targets. Nat Rev Drug Discov 2011;10:903-14. [PubMed]
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007;9:166-80. [Crossref] [PubMed]
- Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004;6:1-6. [Crossref] [PubMed]
- MacDonald JW, Ghosh D. COPA--cancer outlier profile analysis. Bioinformatics 2006;22:2950-1. [Crossref] [PubMed]
- Wilson BJ, Giguère V. Identification of novel pathway partners of p68 and p72 RNA helicases through Oncomine meta-analysis. BMC Genomics 2007;8:419. [Crossref] [PubMed]
- Blonska M, Lin X. NF-κB signaling pathways regulated by CARMA family of scaffold proteins. Cell Res 2011;21:55-70. [Crossref] [PubMed]
- McDaniel AS, Hovelson D, Cani A, et al. 187 Targeted genomic profiling of penile squamous cell carcinoma using the Oncomine cancer research panel. Eur J Cancer 2014;50:61. [Crossref]
- Wang CY, Lai MD, Phan NN, et al. Meta-Analysis of Public Microarray Datasets Reveals Voltage-Gated Calcium Gene Signatures in Clinical Cancer Patients. PLoS One 2015;10:e0125766 [Crossref] [PubMed]
- Phan NN, Wang CY, Chen CF, et al. Voltage-gated calcium channels: Novel targets for cancer therapy. Oncol Lett 2017;14:2059-74. [PubMed]
- Wang CY, Shahi P, Huang JT, et al. Systematic analysis of the achaete-scute complex-like gene signature in clinical cancer patients. Mol Clin Oncol 2017;6:7-18. [Crossref] [PubMed]
- Coppola G. editor. The OMICs: Applications in Neuroscience. Oxford: Oxford University Press, 2013.
- Cohen T, Sundaresh S, Levine F. Antipsychotics activate the TGFβ pathway effector SMAD3. Mol Psychiatry 2013;18:347-57. [Crossref] [PubMed]
- Kupershmidt I, Su QJ, Grewal A, et al. Ontology-based meta-analysis of global collections of high-throughput public data. PLoS One 2010;5: [Crossref] [PubMed]
- Ewald JA, Downs TM, Cetnar JP, et al. Expression microarray meta-analysis identifies genes associated with Ras/MAPK and related pathways in progression of muscle-invasive bladder transition cell carcinoma. PLoS One 2013;8:e55414 [Crossref] [PubMed]
- Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 2009;6:e1000097 [Crossref] [PubMed]
- Rhodes DR, Chinnaiyan AM. Integrative analysis of the cancer transcriptome. Nat Genet 2005;37:S31-7. [Crossref] [PubMed]
- Győrffy B, Surowiak P, Budczies J, et al. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One 2013;8:e82241 [Crossref] [PubMed]
- Goswami CP, Nakshatri H. PROGgeneV2: enhancements on the existing database. BMC Cancer 2014;14:970. [Crossref] [PubMed]
- Momin A, Cadiou H, Mason A, et al. Role of the hyperpolarization-activated current Ih in somatosensory neurons. J Physiol 2008;586:5911-29. [Crossref] [PubMed]
- Alonso EN, Orozco M, Eloy Nieto A, et al. Genes related to suppression of malignant phenotype induced by Maitake D-Fraction in breast cancer cells. J Med Food 2013;16:602-17. [Crossref] [PubMed]
- Kuo SH, Yang SY, You SL, et al. Polymorphisms of ESR1, UGT1A1, HCN1, MAP3K1 and CYP2B6 are associated with the prognosis of hormone receptor-positive early breast cancer. Oncotarget 2017;8:20925-38. [PubMed]
- Lau YT, Wong CK, Luo J, et al. Effects of hyperpolarization-activated cyclic nucleotide-gated (HCN) channel blockers on the proliferation and cell cycle progression of embryonic stem cells. Pflugers Arch 2011;461:191-202. [Crossref] [PubMed]
- Poolos NP. The Yin and Yang of the H-Channel and Its Role in Epilepsy. Epilepsy Curr 2004;4:3-6. [Crossref] [PubMed]
- Ueda K, Nakamura K, Hayashi T, et al. Functional characterization of a trafficking-defective HCN4 mutation, D553N, associated with cardiac arrhythmia. J Biol Chem 2004;279:27194-8. [Crossref] [PubMed]
- Emery EC, Young GT, Berrocoso EM, et al. HCN2 ion channels play a central role in inflammatory and neuropathic pain. Science 2011;333:1462-6. [Crossref] [PubMed]
- Yu X, Duan KL, Shang CF, et al. Calcium influx through hyperpolarization-activated cation channels (Ih channels) contributes to activity-evoked neuronal secretion. Proc Natl Acad Sci U S A 2004;101:1051-6. [Crossref] [PubMed]
- Norberg E, Karlsson M, Korenovska O, et al. Critical role for hyperpolarization-activated cyclic nucleotide-gated channel 2 in the AIF-mediated apoptosis. EMBO J 2010;29:3869-78. [Crossref] [PubMed]
- Ulens C, Tytgat J. Gi- and Gs-coupled receptors up-regulate the cAMP cascade to modulate HCN2, but not HCN1 pacemaker channels. Pflugers Arch 2001;442:928-42. [Crossref] [PubMed]
- Wainger BJ, DeGennaro M, Santoro B, et al. Molecular mechanism of cAMP modulation of HCN pacemaker channels. Nature 2001;411:805-10. [Crossref] [PubMed]
- Norberg E, Gogvadze V, Ott M, et al. An increase in intracellular Ca2+ is required for the activation of mitochondrial calpain to release AIF during cell death. Cell Death Differ 2008;15:1857-64. [Crossref] [PubMed]
- Liu Z, Xie M, Yao Z, et al. Three meta-analyses define a set of commonly overexpressed genes from microarray datasets on astrocytomas. Mol Neurobiol 2013;47:325-36. [Crossref] [PubMed]
- Cai Y, Zhong X, Wang Y, et al. Screening feature genes of astrocytoma using a combined method of microarray gene expression profiling and bioinformatics analysis. Int J Clin Exp Med 2015;8:18004. [PubMed]
- Etcheverry A, Aubry M, De Tayrac M, et al. DNA methylation in glioblastoma: impact on gene expression and clinical outcome. BMC Genomics 2010;11:701. [Crossref] [PubMed]
- Sun L, Hui AM, Su Q, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell 2006;9:287-300. [Crossref] [PubMed]
- Liu S, Yin F, Zhang J, et al. Regulatory roles of miRNA in the human neural stem cell transformation to glioma stem cells. J Cell Biochem 2014;115:1368-80. [Crossref] [PubMed]
- Madhavan S, Zenklusen JC, Kotliarov Y, et al. Rembrandt: helping personalized medicine become a reality through integrative translational research. Mol Cancer Res 2009;7:157-67. [Crossref] [PubMed]
- Liu Z, Yao Z, Li C, et al. Gene expression profiling in human high-grade astrocytomas. Comp Funct Genomics 2011;2011:245137 [Crossref] [PubMed]
- Zhao X, Liu Z, Yu L, et al. Global gene expression profiling confirms the molecular fidelity of primary tumor-based orthotopic xenograft mouse models of medulloblastoma. Neuro Oncol 2012;14:574-83. [Crossref] [PubMed]
- Pollard SM, Yoshikawa K, Clarke ID, et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell stem cell 2009;4:568-80. [Crossref] [PubMed]
- Tayrac Md, Etcheverry A, Aubry M, et al. Integrative genome-wide analysis reveals a robust genomic glioblastoma signature associated with copy number driving changes in gene expression. Genes Chromosomes Cancer 2009;48:55-68. [Crossref] [PubMed]
- Liang Y, Diehn M, Watson N, et al. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proc Natl Acad Sci U S A 2005;102:5814-9. [Crossref] [PubMed]
- McLendon R, Friedman A, Bigner D, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008;455:1061-8. [Crossref] [PubMed]
- Griesinger AM, Birks DK, Donson AM, et al. Characterization of distinct immunophenotypes across pediatric brain tumor types. J Immunol 2013;191:4880-8. [Crossref] [PubMed]
- Birks DK, Donson AM, Patel PR, et al. Pediatric rhabdoid tumors of kidney and brain show many differences in gene expression but share dysregulation of cell cycle and epigenetic effector genes. Pediatr Blood Cancer 2013;60:1095-102. [Crossref] [PubMed]
- Valdora F, Banelli B, Stigliani S, et al. Epigenetic silencing of DKK3 in medulloblastoma. Int J Mol Sci 2013;14:7492-505. [Crossref] [PubMed]
- Lambert SR, Witt H, Hovestadt V, et al. Differential expression and methylation of brain developmental genes define location-specific subsets of pilocytic astrocytoma. Acta Neuropathol 2013;126:291-301. [Crossref] [PubMed]
- Raskin L, Fullen DR, Giordano TJ, et al. Transcriptome profiling identifies HMGA2 as a biomarker of melanoma progression and prognosis. J Invest Dermatol 2013;133:2585-92. [Crossref] [PubMed]
- Łastowska M, Viprey V, Santibanez-Koref M, et al. Identification of candidate genes involved in neuroblastoma progression by combining genomic and expression microarrays with survival data. Oncogene 2007;26:7432-44. [Crossref] [PubMed]
- Jessen WJ, Miller SJ, Jousma E, et al. MEK inhibition exhibits efficacy in human and mouse neurofibromatosis tumors. J Clin Invest 2013;123:340-7. [Crossref] [PubMed]
- Miller SJ, Jessen WJ, Mehta T, et al. Integrative genomic analyses of neurofibromatosis tumours identify SOX9 as a biomarker and survival gene. EMBO Mol Med 2009;1:236-48. [Crossref] [PubMed]
- Harms PW, Patel RM, Verhaegen ME, et al. Distinct gene expression profiles of viral-and nonviral-associated merkel cell carcinoma revealed by transcriptome analysis. J Invest Dermatol 2013;133:936-45. [Crossref] [PubMed]
- Chen X, Leung SY, Yuen ST, et al. Variation in gene expression patterns in human gastric cancers. Mol Biol Cell 2003;14:3208-15. [Crossref] [PubMed]
- Neumann O, Kesselmeier M, Geffers R, et al. Methylome analysis and integrative profiling of human HCCs identify novel protumorigenic factors. Hepatology 2012;56:1817-27. [Crossref] [PubMed]
- Sia D, Hoshida Y, Villanueva A, et al. Integrative molecular analysis of intrahepatic cholangiocarcinoma reveals 2 classes that have different outcomes. Gastroenterology 2013;144:829-40. [Crossref] [PubMed]
- Jerez A, Clemente MJ, Makishima H, et al. STAT3 mutations unify the pathogenesis of chronic lymphoproliferative disorders of NK cells and T-cell large granular lymphocyte leukemia. Blood 2012;120:3048-57. [Crossref] [PubMed]
- April C, Klotzle B, Royce T, et al. Whole-genome gene expression profiling of formalin-fixed, paraffin-embedded tissue samples. PLoS One 2009;4:e8162 [Crossref] [PubMed]
- Frampton AE, Castellano L, Colombo T, et al. MicroRNAs cooperatively inhibit a network of tumor suppressor genes to promote pancreatic tumor growth and progression. Gastroenterology 2014;146:268-77.e18. [Crossref] [PubMed]
- Bredel M, Bredel C, Juric D, et al. Functional network analysis reveals extended gliomagenesis pathway maps and three novel MYC-interacting genes in human gliomas. Cancer Res 2005;65:8679-89. [Crossref] [PubMed]
- Murat A, Migliavacca E, Gorlia T, et al. Stem cell–related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol 2008;26:3015-24. [Crossref] [PubMed]
- Glück S, Ross JS, Royce M, et al. TP53 genomics predict higher clinical and pathologic tumor response in operable early-stage breast cancer treated with docetaxel-capecitabine±trastuzumab. Breast Cancer Res Treat 2012;132:781-91. [Crossref] [PubMed]
- Storz MN, van de Rijn M, Kim YH, et al. Gene expression profiles of cutaneous B cell lymphoma. J Invest Dermatol 2003;120:865-70. [Crossref] [PubMed]
- Piccaluga PP, Agostinelli C, Califano A, et al. Gene expression analysis of peripheral T cell lymphoma, unspecified, reveals distinct profiles and new potential therapeutic targets. J Clin Invest 2007;117:823. [Crossref] [PubMed]
- Riker AI, Enkemann SA, Fodstad O, et al. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med Genomics 2008;1:13. [Crossref] [PubMed]
- Huang Q, Lin B, Liu H, et al. RNA-Seq analyses generate comprehensive transcriptomic landscape and reveal complex transcript patterns in hepatocellular carcinoma. PLoS One 2011;6:e26168 [Crossref] [PubMed]
- Sperger JM, Chen X, Draper JS, et al. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc Natl Acad Sci U S A 2003;100:13350-5. [Crossref] [PubMed]
- Costa V, Esposito R, Ziviello C, et al. New somatic mutations and WNK1-B4GALNT3 gene fusion in papillary thyroid carcinoma. Oncotarget 2015;6:11242. [Crossref] [PubMed]
- Li L, Wei Y, To C, et al. Integrated omic analysis of lung cancer reveals metabolism proteome signatures with prognostic impact. Nat Commun 2014;5:5469. [Crossref] [PubMed]
- Kort EJ, Farber L, Tretiakova M, et al. The E2F3-Oncomir-1 axis is activated in Wilms' tumor. Cancer Res 2008;68:4034-8. [Crossref] [PubMed]
- Hu K, Yu J, Suknuntha K, et al. Efficient generation of transgene-free induced pluripotent stem cells from normal and neoplastic bone marrow and cord blood mononuclear cells. Blood 2011;117:e109-19. [Crossref] [PubMed]
- Park M, Kim M, Hwang D, et al. Characterization of gene expression and activated signaling pathways in solid-pseudopapillary neoplasm of pancreas. Mod Pathol 2014;27:580-93. [Crossref] [PubMed]
- Ooi A, Wong JC, Petillo D, et al. An antioxidant response phenotype shared between hereditary and sporadic type 2 papillary renal cell carcinoma. Cancer Cell 2011;20:511-23. [Crossref] [PubMed]
- Hoek K, Rimm DL, Williams KR, et al. Expression profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer Res 2004;64:5270-82. [Crossref] [PubMed]
- Eckerle S, Brune V, Döring C, et al. Gene expression profiling of isolated tumour cells from anaplastic large cell lymphomas: insights into its cellular origin, pathogenesis and relation to Hodgkin lymphoma. Leukemia 2009;23:2129-38. [Crossref] [PubMed]
- Brune V, Tiacci E, Pfeil I, et al. Origin and pathogenesis of nodular lymphocyte–predominant Hodgkin lymphoma as revealed by global gene expression analysis. J Exp Med 2008;205:2251-68. [Crossref] [PubMed]
- Mailloux AW, Zhang L, Moscinski L, et al. Fibrosis and Subsequent Cytopenias Are Associated with Basic Fibroblast Growth Factor–Deficient Pluripotent Mesenchymal Stromal Cells in Large Granular Lymphocyte Leukemia. J Immunol 2013;191:3578-93. [Crossref] [PubMed]
- Lapointe J, Li C, Higgins JP, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci U S A 2004;101:811-6. [Crossref] [PubMed]
- Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 2009;27:1160-7. [Crossref] [PubMed]
- Crnogorac-Jurcevic T, Chelala C, Barry S, et al. Molecular analysis of precursor lesions in familial pancreatic cancer. PLoS One 2013;8:e54830 [Crossref] [PubMed]
- Hu Z, Fan C, Livasy C, et al. A compact VEGF signature associated with distant metastases and poor outcomes. BMC Med 2009;7:9. [Crossref] [PubMed]
- De Preter K, Vandesompele J, Heimann P, et al. Human fetal neuroblast and neuroblastoma transcriptome analysis confirms neuroblast origin and highlights neuroblastoma candidate genes. Genome Biol 2006;7:R84. [Crossref] [PubMed]
- Uva P, Aurisicchio L, Watters J, et al. Comparative expression pathway analysis of human and canine mammary tumors. BMC Genomics 2009;10:135. [Crossref] [PubMed]
- Shimokuni T, Tanimoto K, Hiyama K, et al. Chemosensitivity prediction in esophageal squamous cell carcinoma: novel marker genes and efficacy-prediction formulae using their expression data. Int J Oncol 2006;28:1153-62. [PubMed]
- Joy A, Ramesh A, Smirnov I, et al. AKT pathway genes define 5 prognostic subgroups in glioblastoma. PLoS One 2014;9:e100827 [Crossref] [PubMed]
- Griesinger AM, Josephson RJ, Donson AM, et al. Interleukin-6/STAT3 pathway signaling drives an inflammatory phenotype in Group A ependymoma. Cancer Immunol Res 2015;3:1165-74. [Crossref] [PubMed]
- Shah MV, Zhang R, Irby R, et al. Molecular profiling of LGL leukemia reveals role of sphingolipid signaling in survival of cytotoxic lymphocytes. Blood 2008;112:770-81. [Crossref] [PubMed]
- Yuan SX, Wang J, Yang F, et al. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology 2016;63:499-511. [Crossref] [PubMed]
- Tong M, Chan KW, Bao JY, et al. Rab25 is a tumor suppressor gene with antiangiogenic and anti-invasive activities in esophageal squamous cell carcinoma. Cancer Res 2012;72:6024-35. [Crossref] [PubMed]
- Wei G, Luo H, Sun Y, et al. Transcriptome profiling of esophageal squamous cell carcinoma reveals a long noncoding RNA acting as a tumor suppressor. Oncotarget 2015;6:17065. [Crossref] [PubMed]
- Gutmann DH, Hedrick NM, Li J, et al. Comparative gene expression profile analysis of neurofibromatosis 1-associated and sporadic pilocytic astrocytomas. Cancer Res 2002;62:2085-91. [PubMed]
- Shai R, Shi T, Kremen TJ, et al. Gene expression profiling identifies molecular subtypes of gliomas. Oncogene 2003;22:4918-23. [Crossref] [PubMed]
- French PJ, Swagemakers SM, Nagel JH, et al. Gene expression profiles associated with treatment response in oligodendrogliomas. Cancer Res 2005;65:11335-44. [Crossref] [PubMed]
- Kaiser S, Park YK, Franklin JL, et al. Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome Biol 2007;8:R131. [Crossref] [PubMed]
- Jones J, Otu H, Spentzos D, et al. Gene signatures of progression and metastasis in renal cell cancer. Clin Cancer Res 2005;11:5730-9. [Crossref] [PubMed]
- Yusenko MV, Kuiper RP, Boethe T, et al. High-resolution DNA copy number and gene expression analyses distinguish chromophobe renal cell carcinomas and renal oncocytomas. BMC Cancer 2009;9:152. [Crossref] [PubMed]
- Cutcliffe C, Kersey D, Huang CC, et al. Clear cell sarcoma of the kidney: up-regulation of neural markers with activation of the sonic hedgehog and Akt pathways. Clin Cancer Res 2005;11:7986-94. [Crossref] [PubMed]
- Basso K, Margolin AA, Stolovitzky G, et al. Reverse engineering of regulatory networks in human B cells. Nat Genet 2005;37:382-90. [Crossref] [PubMed]
- Valk PJ, Verhaak RG, Beijen MA, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med 2004;350:1617-28. [Crossref] [PubMed]
- Dürig J, Bug S, Klein-Hitpass L, et al. Combined single nucleotide polymorphism-based genomic mapping and global gene expression profiling identifies novel chromosomal imbalances, mechanisms and candidate genes important in the pathogenesis of T-cell prolymphocytic leukemia with inv (14)(q11q32). Leukemia 2007;21:2153-63. [Crossref] [PubMed]
- Bhattacharjee A, Richards WG, Staunton J, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A 2001;98:13790-5. [Crossref] [PubMed]
- Talantov D, Mazumder A, Jack XY, et al. Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin Cancer Res 2005;11:7234-42. [Crossref] [PubMed]
- Grützmann R, Pilarsky C, Ammerpohl O, et al. Gene expression profiling of microdissected pancreatic ductal carcinomas using high-density DNA microarrays. Neoplasia 2004;6:611-22. [Crossref] [PubMed]
- Buchholz M, Braun M, Heidenblut A, et al. Transcriptome analysis of microdissected pancreatic intraepithelial neoplastic lesions. Oncogene 2005;24:6626-36. [Crossref] [PubMed]
- Detwiller KY, Fernando NT, Segal NH, et al. Analysis of hypoxia-related gene expression in sarcomas and effect of hypoxia on RNA interference of vascular endothelial cell growth factor A. Cancer Res 2005;65:5881-9. [Crossref] [PubMed]
- Henriquez NV, Forshew T, Tatevossian R, et al. Comparative expression analysis reveals lineage relationships between human and murine gliomas and a dominance of glial signatures during tumor propagation in vitro. Cancer Res 2013;73:5834-44. [Crossref] [PubMed]
- Romero A, Martín M, Oliva B, et al. Glutathione S-transferase P1 c.313A > G polymorphism could be useful in the prediction of doxorubicin response in breast cancer patients. Ann Oncol 2012;23:1750-6. [Crossref] [PubMed]
- Stathopoulos GP, Armakolas A. Differences in gene expression between individuals with multiple primary and single primary malignancies. Int J Mol Med 2009;24:613-22. [Crossref] [PubMed]
- Kim WJ, Kim EJ, Kim SK, et al. Predictive value of progression-related gene classifier in primary non-muscle invasive bladder cancer. Mol Cancer 2010;9:3. [Crossref] [PubMed]
- Kim YJ, Yoon HY, Kim JS, et al. HOXA9, ISL1 and ALDH1A3 methylation patterns as prognostic markers for nonmuscle invasive bladder cancer: Array-based DNA methylation and expression profiling. Int J Cancer 2013;133:1135-42. [Crossref] [PubMed]
- Abba M, Laufs S, Aghajany M, et al. Look who’s talking: deregulated signaling in colorectal cancer. Cancer Genomics Proteomics 2012;9:15-25. [PubMed]
- Ema A, Waraya M, Yamashita K, et al. Identification of EGFR expression status association with metastatic lymph node density (ND) by expression microarray analysis of advanced gastric cancer. Cancer Med 2015;4:90-100. [Crossref] [PubMed]
- Yang C, Zhuang Z, Fliedner SM, et al. Germ-line PHD1 and PHD2 mutations detected in patients with pheochromocytoma/paraganglioma-polycythemia. J Mol Med 2015;93:93-104. [Crossref] [PubMed]
- Gao F, Liang H, Lu H, et al. Global analysis of DNA methylation in hepatocellular carcinoma by a liquid hybridization capture-based bisulfite sequencing approach. Clin Epigenetics 2015;7:86. [Crossref] [PubMed]
- Leja J, Essaghir A, Essand M, et al. Novel markers for enterochromaffin cells and gastrointestinal neuroendocrine carcinomas. Mod Pathol 2009;22:261-72. [Crossref] [PubMed]
- Ding X, Yang Y, Han B, et al. Transcriptomic characterization of hepatocellular carcinoma with CTNNB1 mutation. PLoS One 2014;9:e95307 [Crossref] [PubMed]
- Kadara H, Fujimoto J, Yoo SY, et al. Transcriptomic architecture of the adjacent airway field cancerization in non–small cell lung cancer. J Natl Cancer Inst 2014;106:dju004 [Crossref] [PubMed]
- Rousseaux S, Debernardi A, Jacquiau B, et al. Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers. Sci Transl Med 2013;5:186ra66 [Crossref] [PubMed]
- Lockwood WW, Chari R, Coe BP, et al. DNA amplification is a ubiquitous mechanism of oncogene activation in lung and other cancers. Oncogene 2008;27:4615-24. [Crossref] [PubMed]
- Smith AP, Hoek K, Becker D. Whole-genome expression profiling of the melanoma progression pathway reveals marked molecular differences between nevi/melanoma in situ and advanced-stage melanomas. Cancer Biol Ther 2005;4:1018-29. [Crossref] [PubMed]
- Chandran UR, Ma C, Dhir R, et al. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer 2007;7:64. [Crossref] [PubMed]
- Renner M, Wolf T, Meyer H, et al. Integrative DNA methylation and gene expression analysis in high-grade soft tissue sarcomas. Genome Biol 2013;14:r137. [Crossref] [PubMed]
- Grasso CS, Wu YM, Robinson DR, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012;487:239-43. [Crossref] [PubMed]
- Sia D, Losic B, Moeini A, et al. Massive parallel sequencing uncovers actionable FGFR2-PPHLN1 fusion and ARAF mutations in intrahepatic cholangiocarcinoma. Nat Commun 2015;6:6087. [Crossref] [PubMed]
- Aoyagi K, Minashi K, Igaki H, et al. Artificially induced epithelial-mesenchymal transition in surgical subjects: its implications in clinical and basic cancer research. PLoS One 2011;6:e18196 [Crossref] [PubMed]
- Yu K, Ganesan K, Tan LK, et al. A precisely regulated gene expression cassette potently modulates metastasis and survival in multiple solid cancers. PLoS Genet 2008;4:e1000129 [Crossref] [PubMed]
- Sternberg A, Killick S, Littlewood T, et al. Evidence for reduced B-cell progenitors in early (low-risk) myelodysplastic syndrome. Blood 2005;106:2982-91. [Crossref] [PubMed]
- LaBreche HG, Nevins JR, Huang E. Integrating factor analysis and a transgenic mouse model to reveal a peripheral blood predictor of breast tumors. BMC Med Genomics 2011;4:61. [Crossref] [PubMed]
- Seok JY, Na DC, Woo HG, et al. A fibrous stromal component in hepatocellular carcinoma reveals a cholangiocarcinoma-like gene expression trait and epithelial-mesenchymal transition. Hepatology 2012;55:1776-86. [Crossref] [PubMed]
- Zhan F, Barlogie B, Arzoumanian V, et al. Gene-expression signature of benign monoclonal gammopathy evident in multiple myeloma is linked to good prognosis. Blood 2007;109:1692-700. [Crossref] [PubMed]
- Gordon GJ, Rockwell GN, Jensen RV, et al. Identification of novel candidate oncogenes and tumor suppressors in malignant pleural mesothelioma using large-scale transcriptional profiling. Am J Pathol 2005;166:1827-40. [Crossref] [PubMed]
- Wu H, Chen Y, Liang J, et al. Hypomethylation-linked activation of PAX2 mediates tamoxifen-stimulated endometrial carcinogenesis. Nature 2005;438:981-7. [Crossref] [PubMed]
- Dyrskjøt L, Kruhøffer M, Thykjaer T, et al. Gene Expression in the Urinary Bladder A Common Carcinoma in Situ Gene Expression Signature Exists Disregarding Histopathological Classification. Cancer Res 2004;64:4040-8. [Crossref] [PubMed]
- Radvanyi L, Singh-Sandhu D, Gallichan S, et al. The gene associated with trichorhinophalangeal syndrome in humans is overexpressed in breast cancer. Proc Natl Acad Sci U S A 2005;102:11005-10. [Crossref] [PubMed]
- Kimchi ET, Posner MC, Park JO, et al. Progression of Barrett's metaplasia to adenocarcinoma is associated with the suppression of the transcriptional programs of epidermal differentiation. Cancer Res 2005;65:3146-54. [Crossref] [PubMed]
- Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. J Clin Epidemiol 2009;62:1006-12. [Crossref] [PubMed]


