
Vesicular control of metastasis: entrap the “virtuous” and free the “wicked” microRNA
Metastasis, the process by which cancers disseminate to sites distant from the primary tumor, is a complex process orchestrated by genetic, epigenetic and microenvironmental factors (1). This process is initiated and sustained by an accelerated Darwinian-based selection of cellular clones capable of invading the extracellular matrix, penetrating veins and lymphatic vessels (intravasation), surviving a hostile circulation environment, extravasation, arresting at distant organs, and ultimately colonizing them (2). The process is extraordinarily inefficient, yet accounts for more than 90% of cancer mortality. Thus, our ability to understand the process is imperative for predicting cancer behavior, its prognosis, diagnostic biomarkers and ultimately successful treatment. However, despite the recent advances in understanding the cellular and molecular basis of metastasis, effective treatment options are lacking and the molecular network pathologies underpinning its intricacies remain elusive. Deciphering the genetic basis of cancer metastasis has entered a remarkable stage of progress, especially with the advent of next generation sequencing technology. Panoply of evidence suggest that at the DNA level, both chromosomal instability and somatic gene mutations play crucial roles in cancer progression (3). However, the fact that primary tumors and metastases, at least in some cancers, have comparable genetic aberrations suggests more intricate metastasis-promoting mechanisms that may involve aberrant transcriptional and epigenetic processes (4,5). Moreover, it is becoming evident, after the completion of the human genome project, that genome and transcriptome sequencing are not sufficient to predict the complexity of the metastatic process. It is logical for the scientific community to step back and re-examine the intricacies of the metastatic interactome insightfully. There is no better example to illustrate such intricacies than non-coding RNA (ncRNA). Non-coding RNAs, which include microRNA (miRNA), small inhibitory RNAs (siRNA), circular RNAs, piwi interacting RNAs and lncRNAs, have been shown to play significant roles in cancer metastasis (6,7). miRNAs are non-protein coding, 17
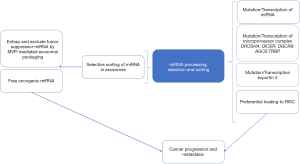
By generating a stably transfected colon cancer cell line expressing a luciferase protein fused to a biotin acceptor peptide with a transmembrane domain of platelet-derived growth factor capable of localizing the chimera in the plasma membranes of extracellular vesicles (ECV), Teng et al. were able to isolate exosomes from the stable colon cancer cell line (CT26) in vitro using streptavidin-coated beads allowing for their miRNA content to be successfully profiled using miRNA microarray (9). Similarly, transplanting the stable donor CT26 in the colonic submucosa or in the spleen of BALB/c mice, allowed the authors to track and compare ECV miRNA contents in primary tumors and their liver metastases and separate them from miRNAs harbored in non-cancer-derived exosomes. Having established this “witty” model, the authors showed elegantly significant differences in ECV packaged miRNA derived from primary CT26 tumors, metastases and normal colonic mucosa compared to each other and their original donor cells, hinting at an active miRNA sorting process in ECV. For example, while only 25.8% of miRNAs were equally shared between primary tumors and donor cells, 47.5% of miRNAs were lower and 26.7% were higher in primary tumor-derived exosomes compared to exosomal donor cells. Similarly, CT26 cells that metastasized to the liver had significantly higher miRNA-193a and lower miRNA 18a and miRNA 21 expression in ECV compared to primary colon cancer derived exosomes while some of the exosomal miRNAs retained the same pattern. Interestingly, in metastatic tissues miRNA 193a was lower and miRNA 21 was higher, indicating a selective sorting of miRNA 193a, among others, in exosomes. Functional classification of miRNAs based on their oncogenic or suppressive nature revealed a striking depiction in that most of the miRNAs with tumor suppressive activity (miRNA 10a-5p, miRNA 193a-3p, miRNA 200b-5p and miRNA 222-3p) were actively packaged into exosomes and released in the circulation where they can be detected in plasma and urine using quantitative PCR methodologies. Conversely, oncogenic miRNAs, like miRNA 21, levels were higher in primary tumor tissues and liver metastases than in their exosomes. The data of Teng et al., clearly suggest selective sorting mechanisms of miRNAs in cancer cells mediated by exosomes that ultimately potentiate cancer progression. One wonders, whether in cancers with low metastatic potential, like basal cell carcinoma, the opposite selection of miRNA would be the norm. Evidently, the data presented by Teng et al., have direct potential relevance to translational medicine in many ways; Measuring the tumor suppressive miRNAs level in exosomes from plasma and urine may be used as prognostic biomarkers in patients with colorectal cancer. Indeed, by assessing levels of three tumor suppressor miRNAs, namely miRNA 193a, miRNA126 and miRNA 148a and the oncogenic miRNA 196b in exosomes from the plasma of patients with metastatic versus non-metastatic colorectal cancer using quantitative PCR, Teng et al., have predictably revealed a significant increase in the three suppressive miRNAs and a reduction of miRNA 196b in metastatic colorectal cancer compared to more indolent tumors. The results potentially illuminate a feasible method for early intervention in patients who are at risk from developing metastatic disease, an avenue that is ironically poorly explored.
How is the selective sorting of miRNA achieved in cells? Using Biotin-tagged miRNA 193a and pull-down from whole cell extract by streptavidin beads and the subsequent separation of the protein bands on 10% SDS-PAGE, followed by in gel digestion of differential bands and MALDI-TOF mass spectrometry, the authors identified major vault protein (MVP) as a dose-dependent binding partner of miRNA 193a. It has been previously established that MVP, a ribonucleoprotein, is overexpressed in multi-drug resistant cancers including gliomas, ovarian and breast cancer. MVP has also been shown to act as a substrate for the tyrosine phosphatase SHP-2 and scaffold protein in epidermal growth factor signaling and in the negative regulation of c-Jun-mediated activator protein 1 transcription in mammalian cells (10). Further confirmatory experiments utilizing CRISPR knockout of MVP and its overexpression established that miR-193a sorting into exosomes is MVP-dependent. Interestingly, MVP expression in human colorectal cancer was stage dependent with stage III cancers involving lymph nodes metastasis displaying the highest levels of MVP expression compared to lower stages. The knockout of MVP in CT26 cells, although resulted in the selective accumulation of miRNA 193a, did not influence other suppressive miRNAs. This important piece of the puzzle suggests that there may be other proteins involved in selective packaging of various miRNAs. With about 2,588 miRNA genes in the human genome reported so far, the list of miRNA sorting proteins, most likely, will continue to expand.
Although the interesting findings of Teng et al., focused mainly on colorectal cancer and its progression, an important issue to be addressed by the scientific community is whether miRNAs’ selective sorting is involved in other cancers or other chronic disorders. Several miRNA therapeutics have reach clinical trial stage. If Teng et al.’s miRNA sorting mechanism proves to be accurate and widespread, the use of external miRNAs as therapeutic agents may become more challenging. Conversely, the future identification and targeting of MVP-like proteins will open other therapeutic avenues against cancer and perhaps other diseases.
Acknowledgments
The author thanks Manar Altahnon and Malak Qaddoumi for their comments on the article.
Funding: F Al-Mulla is funded by Kuwait Foundation for the Advancement of Sciences (KFAS) (grant number: 2011-1302-06).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Gang Wang (Department of Pancreatic and Biliary Surgery, The First Affiliated Hospital of Harbin Medical University, Harbin, China).
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.06.49). The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med 2006;12:895-904. [Crossref] [PubMed]
- Turajlic S, Swanton C. Metastasis as an evolutionary process. Science 2016;352:169-75. [Crossref] [PubMed]
- Ali H, Bitar MS, Al Madhoun A, et al. Functionally-focused algorithmic analysis of high resolution microarray-CGH genomic landscapes demonstrates comparable genomic copy number aberrations in MSI and MSS sporadic colorectal cancer. PLoS One 2017;12:e0171690 [Crossref] [PubMed]
- Al-Mulla F, Keith WN, Pickford IR, et al. Comparative genomic hybridization analysis of primary colorectal carcinomas and their synchronous metastases. Genes Chromosomes Cancer 1999;24:306-14. [Crossref] [PubMed]
- Al-Temaimi RA, Tan TZ, Marafie MJ, et al. Identification of 42 Genes Linked to Stage II Colorectal Cancer Metastatic Relapse. Int J Mol Sci 2016;17:E598 [Crossref] [PubMed]
- Schwarzenbach H. The clinical relevance of circulating, exosomal miRNAs as biomarkers for cancer. Expert Rev Mol Diagn 2015;15:1159-69. [Crossref] [PubMed]
- Zhao M, Ang L, Huang J, et al. MicroRNAs regulate the epithelial-mesenchymal transition and influence breast cancer invasion and metastasis. Tumour Biol 2017;39:1010428317691682 [Crossref] [PubMed]
- Lagos-Quintana M, Rauhut R, Lendeckel W, et al. Identification of novel genes coding for small expressed RNAs. Science 2001;294:853-8. [Crossref] [PubMed]
- Teng Y, Ren Y, Hu X, et al. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nat Commun 2017;8:14448. [Crossref] [PubMed]
- Lötsch D, Steiner E, Holzmann K, et al. Major vault protein supports glioblastoma survival and migration by upregulating the EGFR/PI3K signalling axis. Oncotarget 2013;4:1904-18. [Crossref] [PubMed]


