
Centromere protein F (CENPF) is upregulated and related to tumor differentiation in laryngeal squamous cell carcinoma
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the 6th most common cancer worldwide (1). Laryngeal squamous cell carcinoma (LSCC) is the most common subtype of HNSCC, accounting for more than 90 percent of head and neck cancers (2); subglottic cancer is the most prevalent type of LSCC (3). The incidence of LSCC is approximately 3.5–55.5 per 100,000 males and 0.6 per 100,000 females (4). Although some advances in diagnosis and therapy have been made in recent years, the survival rate for LSCC has not significantly increased (5). Hence, investigation of potential biomarkers for LSCC is essential.
Human centromere protein F (CENPF; also known as mitosin/P330d) was first discovered by Rattner et al. (6). Full-length CENPF is a 367 kDa nucleophosmin, containing 3,210 amino acids (7). CENPF has been associated with various types of cancer, particularly prostate cancer (8). However, there has been no investigation of the role of CENPF in the occurrence and development of LSCC.
This study aimed to investigate the relationship between CENPF and LSCC. The ultimate goal was to clarify whether CENPF could serve as a diagnostic and prognostic biomarker in LSCC.
Methods
Cell culture and main reagents
The HNSCC cell lines FaDu and Hep-2 were cultured in RMPI-1640 (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (HyClone, Logan, UT, USA) at 37 °C in 5% CO2. RPMI-1640 and 0.25% trypsin solution were purchased from Invitrogen. Cells in logarithmic growth phase were used for western blotting.
Gene expression profiling
The GEO datasets GSE6631 (GSE6631 expression data from HNSCC), GSE58911 (gene expression in normal and tumor samples from patients with HNSCC), GSE59102 (HOX genes: potential candidates furthering the development of laryngeal squamous cell carcinoma), GSE39366 [molecular subtypes in head and neck cancer (expression)], and GSE27020 (identification and validation of a multigene predictor of recurrence in primary laryngeal cancer) were used to investigate CENPF expression in LSCC and explore the associations between CENPF expression and tumor differentiation and prognosis.
Tissue microarray (TMA)
A commercial LSCC TMA including 5 normal laryngeal tissues, 5 cancer-adjacent normal laryngeal tissues and 30 LSCC tissues derived from patients with an average age of 55.3 years was obtained from Alina Biotechnology Co., Ltd. (Product No. LP804; Xi’an, China). Detailed information on the tissues in the TMA is presented in Table 1.
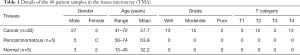
Full table
Immunohistochemical staining
The TMA was deparaffinized in standard pure xylene for 15 min at room temperature 3 times, rehydrated using graded alcohols, and washed in phosphate buffer saline (PBS). Antigen retrieval was performed by boiling in citrate buffer (pH 6.0) for 15 min, then the TMA was cooled to room temperature in the buffer. After washing 3 times in PBS for 5 min, 0.3% hydrogen peroxide phosphate-citrate buffer was applied to block endogenous peroxidase activity for 10 min, then the TMA was washed in PBS for 5 min, incubated with primary CENPF antibody (Sigma-Aldrich; dilution 1:100) for 12 h at 4 °C, followed by poly-HRP goat anti-rabbit (Maixin Bio., FuZhou, China) for 30 min, incubated in diaminobenzidine for 5 min, counterstained using hematoxylin, dehydrated and mounted. Images of the TMA were taken using an Olympus BX40 microscope and CC-12 Soft-Imaging System (Olympus, Tokyo, Japan).
Evaluation of immunohistochemical staining
CENPF demonstrated both cytoplasmic and nuclear expression. CENPF expression in the TMA was scored based on intensity (grade 0, negative; grade 1, weak intensity; grade 2, moderate intensity; grade 3, strong intensity) and frequency (0, 0–25% tumor cells positive; 1, 26–50%; 2, 51–75%; and 3, 76–100%). For statistical analysis, CENPF protein intensity and frequency were transformed into a composite expression score (CES) utilizing the formula CES = intensity × frequency; CES ranged from 0 to 12. The CES was classified as negative [0], weak positive [1–4], positive [5–8], strong positive [9–12].
Silencing of CENPF
ShRNAs molecules used to silence CENPF and a negative control shRNA (which does not target any sequence present in the LSCC cell genome) were obtained from RiboBio Co. Ltd. (Guangzhou, China). LSCC cells were transiently transfected with the shRNAs using Lipofectamine 2000 regent (Invitrogen) according to the manufacturer’s instructions. After silencing, we used flow cytometry for detection of apoptosis. Cells were harvested by trypsinization, washed twice with cold PBS, resuspended in annexin-V-binding buffer and stained with 5 µL of annexin-V/FITC solution and 10 µL of propidium-iodide (PI) solution in the dark for 15 min at room temperature. Fluorescence was analyzed using a FACSCanto™ II spectrometer (BD Biosciences, San Jose, CA, USA). Cells positive for FITC and PI were counted as apoptotic cells. We also conducted cell proliferation assays. Cells were seeded at 1×103 cells/well in 96-well plates in RPMI-1640 medium supplemented with 10% FBS. Twenty-four hours later, the cells were transfected with the shRNAs, and proliferation was determined at 0, 24, 48 and 72 h post-transfection using the Cell Counting Kit-8 (CCK-8) kit (Sigma-Aldrich, Darmstadt, Germany), according to the manufacturer’s instructions.
Western blotting
Total cell lysates were resolved using 10% SDS-PAGE, transferred to PVDF membranes and immunoblotted with primary CENPF antibody (Sigma-Aldrich) and secondary poly-HRP goat anti-rabbit antibody (Maixin Bio, FuZhou, China), following standard protocols. Protein bands were detected using enhanced chemiluminescence reagent (Thermo Fisher Scientific, Rockford, IL, USA).
Statistical analysis
All statistical analyses were performed using Statistical Package for Social Sciences (SPSS) version 20.0 (SPSS, Chicago, USA). Data downloaded from GEO datasets were analyzed using cluster heatmaps. CENPF expression was compared between two groups using t-tests and three or more groups using single factor analysis of variance. Probability values <0.05 were considered statistically significant.
Results
CENPF expression is significantly upregulated in LSCC tissues
We compared the relative expression of CENPF in normal tissues and LSCC using the GEO datasets GSE6631, GSE58911 and GSE59102. CENPF expression was higher in LSCC than normal tissues (Figure 1A, ID: 37302_at, P<0.001; Figure 1B, ID: 7909708, P<0.001; Figure 1C, ID: A_23_P401, P<0.001).
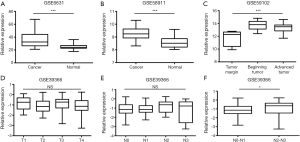
CENPF expression is significantly higher in N2-N3 LSCC compared to N0-N1 LSCC
Analysis of the GSE39366 dataset revealed CENPF expression was not associated with T category (Figure 1D, ID: 18231, P=0.375) or N category (Figure 1E, ID: 18231, P=0.056) in LSCC. However, the expression of CENPF was higher in N2-N3 LSCC than N0-N1 LSCC (Figure 1F, ID: 18231, P=0.029).
CENPF expression is associated with differentiation, prognosis and smoking, but not alcohol consumption
Using the GSE27020 and GSE39366 datasets, we analyzed the associations between CENPF expression and differentiation, prognosis, smoking and alcohol. CENPF was expressed at higher levels in poorly differentiated LSCC than well-differentiated LSCC (Figure 2A, ID: 207828_s_at, P=0.035). Patients with high CENPF expression had poorer overall survival than those with low CENPF expression [Figure 2B, ID: 207828_s_at, HR =2.223 (1.010 to 5.208), P=0.044]. High CENPF expression was associated with smoking (Figure 2C, ID: 18231, P=0.043). However, CENPF expression had no significant association with alcohol consumption (Figure 2D, ID: 18231, P=0.745).

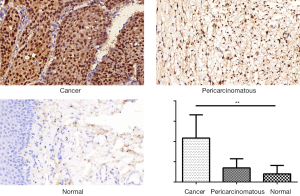
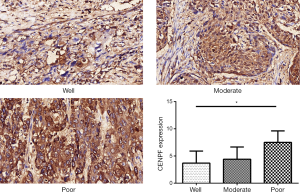
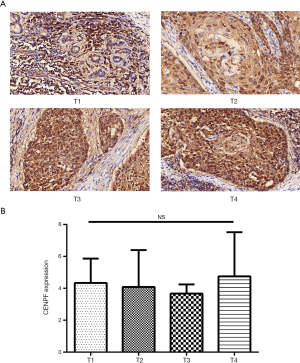
Analysis of CENPF expression in LSCC using the TMA
A TMA was used to analyze the expression of CENPF in clinical LSCC samples. CES scores were determined for every sample in the TMA. CENPF expression was higher in LSCC than normal tissues (Figure 3). Furthermore, CENPF was expressed at higher levels in poorly differentiated tumors than well-differentiated tumors (Figure 4). No association was observed between CENPF and T category (Figure 5).
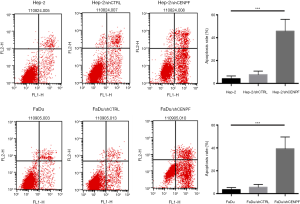
Effect of silencing CENPF in HNSCC cells
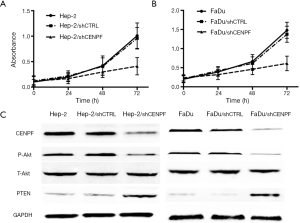
Hep-2 and FaDu cells were transfected with a CENPF shRNA (and named Hep-2/shCENPF and FaDu/shCENPF, respectively). Control cells were transfected with a nonsense shRNA (Hep-2/shCTRL and FaDu/shCTRL). Flow cytometry revealed both Hep-2/shCENPF and FaDu/shCENPF cells contained significantly higher numbers of apoptotic cells than control cells (Figure 6). Silencing CENPF also inhibited the proliferation of Hep-2 (Figure 7A) and FaDu (Figure 7B) cells in a time-dependent manner compared to control cells. Moreover, silencing CENPF reduced the expression of CENPF and markedly altered the levels of proteins related to the PI3K/AKT signaling pathway, including phospho-Akt and PTEN (Figure 7C).
Discussion
Surgery, chemotherapy and radiotherapy are the primary therapeutic modalities for LSCC (9). Though some progress has been achieved, the prognosis of patients with LSCC has not significantly improved (5). Cervical lymph node metastasis is the main factor associated with recurrence and poor prognosis (10). In our clinical practice, approximately one third of patients have lymphatic metastasis when diagnosed with LSCC.
This study investigated the association between CENPF and LSCC. CENPF protein is upregulated in many other malignant neoplasms, including nasopharyngeal cancer and esophageal squamous cell carcinoma (11,12). Both the GEO datasets and TMA revealed that CENPF is significantly upregulated in LSCC.
We also observed CENPF expression was expressed at higher levels in poorly differentiated LSCC compared to well-differentiated LSCC, and in N2-N3 LSCC compared to N0-N1 LSCC. Studies have suggested CENPF drives malignant progression in prostate cancer via a mechanism involving a synergistic interaction between forkhead box M1 (FOXM1) and CENPF (13). Moreover, Lin et al. showed that COUP transcription factor 2 (COUP-TFII) could promote metastasis in prostate cancer through FOXM1 signaling and CENPF (14).
We suggest dysregulation of the miRNA-COUP-TFII-FOXM1-CENPF axis may also occur in LSCC and could promote tumor growth and cervical lymph node metastasis. Analysis of the GSE27020 dataset revealed CENPF was related to differentiation (which was verified by the TMA) and also to the prognosis of patients with LSCC. The results were completely consistent with those in other studies. Though the characteristics and tissue origin of prostate cancer and LSCC vary, CENPF may play a role in development of these tumors via similar mechanisms.
Our results revealed that the tumors of patients who had ever smoked expressed higher levels of CENPF than those who never smoked. Although smoking may promote the occurrence and development of LSCC, there is no consensus on the mechanism (15,16). Liu et al. revealed that oxidative stress is a key factor associated with smoking-related LSCC (17).
We suggest that CENPF also represents a key factor in the development of smoking-related LSCC. In the past 10 years, CENPF has been demonstrated to play important roles in kinetochore-mediated mitotic functions, including maturation of the centromere, chromosome rearrangement and spindle stability (18,19). Therefore, smoking may interfere with mitosis via a mechanism involving CENPF to promote the occurrence of LSCC. In agreement with this suggestion, silencing CENPF promoted apoptosis and inhibited the proliferation of LSCC cells in vitro.
In conclusion, the present study indicates that CENPF may be a potential diagnostic and prognostic biomarker in LSCC, and could represent a potential therapeutic target. Further studies are required to evaluate the value of CENPF as a biomarker and investigate the mechanisms by which it contributes to the development of LSCC.
Study highlights
- What is the current knowledge on the topic—the nucleophosmin CENPF is associated with prostate cancer; its role in the occurrence and development of LSCC is unclear;
- What question did this study address—we investigated the expression of CENPF in LSCC and assessed whether CENPF could be a potential biomarker for LSCC;
- What this study adds to our knowledge—CENPF was upregulated in LSCC compared to normal tissues, and in N2-N3 compared to N0-N1 LSCC. CENPF was associated with differentiation, overall survival and smoking, but not alcohol consumption;
- How this might change clinical pharmacology or translational science—CENPF may represent a diagnostic and prognostic biomarker for LSCC, and may function as a key factor related to smoking-related LSCC.
Acknowledgments
Funding: This work was supported by the Youth Talent Project of Health and Family Planning Commission of Hubei Province (grant number WJ2015Q030); the Wuhan Youth Science and Technology Plan (grant number 2015070404010213) and the Project of Health and Family Planning Commission of Wuhan City (grant number WX13B08).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.08.39). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Vigneswaran N, Williams MD. Epidemiologic trends in head and neck cancer and aids in diagnosis. Oral Maxillofac Surg Clin North Am 2014;26:123-41. [Crossref] [PubMed]
- Ferlay J, Parkin DM, Steliarova-Foucher E. Estimates of cancer incidence and mortality in Europe in 2008. Eur J Cancer 2010;46:765-81. [Crossref] [PubMed]
- Davis RK, Kelly SM, Parkin JL, et al. Selective management of early glottic cancer. Laryngoscope 1990;100:1306-9. [Crossref] [PubMed]
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [Crossref] [PubMed]
- Chin D, Boyle GM, Porceddu S, et al. Head and neck cancer: past, present and future. Expert Rev Anticancer Ther 2006;6:1111-8. [Crossref] [PubMed]
- Rattner JB, Rao A, Fritzler MJ, et al. CENP-F is a.ca 400 kDa kinetochore protein that exhibits a cell-cycle dependent localization. Cell Motil Cytoskeleton 1993;26:214-26. [Crossref] [PubMed]
- Yerushalmi R, Woods R, Ravdin PM, et al. Ki67 in breast cancer: prognostic and predictive potential. Lancet Oncol 2010;11:174-83. [Crossref] [PubMed]
- Lokody I. Signalling: FOXM1 and CENPF: co-pilots driving prostate cancer. Nat Rev Cancer 2014;14:450-1. [Crossref] [PubMed]
- Kaira K, Toyoda M, Shimizu A, et al. Decreasing expression of glucose-regulated protein GRP78/BiP as a significant prognostic predictor in patients with advanced laryngeal squamous cell carcinoma. Head Neck 2016;38:1539-44. [Crossref] [PubMed]
- Xu Y, Zhao X, Guan M, et al. Determination of lymph node micrometastases in patients with supraglottic carcinoma. Acta Otolaryngol 2007;127:1188-95. [Crossref] [PubMed]
- Pimkhaokham A, Shimada Y, Fukuda Y, et al. Nonrandom chromosomal imbalances in esophageal squamous cell carcinoma cell lines: possible involvement of the ATF3 and CENPF genes in the 1q32 amplicon. Jpn J Cancer Res 2000;91:1126-33. [Crossref] [PubMed]
- Cao JY, Liu L, Chen SP, et al. Prognostic significance and therapeutic implications of centromere protein F expression in human nasopharyngeal carcinoma. Mol Cancer 2010;9:237. [Crossref] [PubMed]
- Aytes A, Mitrofanova A, Lefebvre C, et al. Cross-species regulatory network analysis identifies a synergistic interaction between FOXM1 and CENPF that drives prostate cancer malignancy. Cancer Cell 2014;25:638-51. [Crossref] [PubMed]
- Lin SC, Kao CY, Lee HJ, et al. Dysregulation of miRNAs-COUP-TFII-FOXM1-CENPF axis contributes to the metastasis of prostate cancer. Nat Commun 2016;7:11418. [Crossref] [PubMed]
- Bhattacharyya S, Mandal S, Banerjee S, et al. Cannabis smoke can be a major risk factor for early-age laryngeal cancer--a molecular signaling-based approach. Tumour Biol 2015;36:6029-36. [Crossref] [PubMed]
- Shoffel-Havakuk H, Halperin D, Yosef L, et al. The anatomic distribution of malignant and premalignant glottic lesions and its relations to smoking. Otolaryngol Head Neck Surg 2015;152:678-83. [Crossref] [PubMed]
- Liu Y, La C, Wei J, et al. The effect of catalase on smoking related laryngeal squamous cell carcinoma. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi 2015;29:1346-9. [PubMed]
- Coughlin SS, Ekwueme DU. Breast cancer as a global health concern. Cancer Epidemiol 2009;33:315-8. [Crossref] [PubMed]
- Varis A, Salmela AL, Kallio MJ. Cenp-F (mitosin) is more than a mitotic marker. Chromosoma 2006;115:288-95. [Crossref] [PubMed]


