
Role of pharmacogenetics in personalised imatinib dosing
Introduction
Imatinib was the first of the tyrosine kinase inhibitor class of molecularly targeted cancer treatments. It was specifically designed to target the BCR-ABL tyrosine kinase responsible for chronic myeloid leukaemia (CML) pathogenesis, and remains one of the most widely used first-line treatments for CML. Imatinib is also indicated for BCR-ABL-positive acute lymphoblastic leukaemia and, because it also inhibits the c-KIT and PDGFR tyrosine kinases, for c-KIT- and PDGFR-positive gastrointestinal stromal tumours (GISTs), myelodysplastic/myeloproliferative diseases associated with PDGFR gene re-arrangements, aggressive systemic mastocytosis (without D816V c-Kit mutation), hypereosinophilic syndrome and/or chronic eosinophilic leukaemia, and dermatofibrosarcoma protuberans (1-4).
As the indication with the longest history and greatest body of research, and clearest need for personalised dosing, this review will focus on the role of pharmacogenetics in personalised imatinib dosing in the context of CML. However, findings from GIST patients will also be discussed that aid our understanding of genetic contributions to imatinib pharmacokinetics. Furthermore, the conclusions drawn from our experience with imatinib in CML patients should also help inform future imatinib pharmacogenetics research in these other indications.
Then need for personalised imatinib dosing
The arrival of imatinib in 2001 revolutionised CML treatment and dramatically improved patient prognosis compared to the pre-imatinib era. Imatinib dose recommendations for CML treatment have since remained essentially unchanged; a starting dose of 400 mg once daily for all patients, with treatment generally expected to be non-curative and requiring indefinite chronic dosing. However, up to 50% of CML patients will discontinue imatinib due to lack of efficacy or adverse effects when using this one-dose-fits-all approach (5); a significant problem requiring switching to other treatments which may be more costly or may have significant toxicities.
The importance of early treatment response
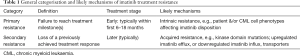
CML treatment efficacy is defined mainly by reduction of BCR-ABL-positive metaphases in bone marrow (cytogenetic response) and reduced BCR-ABL expression in blood cells (molecular response). Standard guidelines identify optimal and suboptimal response, and treatment failure, based on milestones of cytogenetic and molecular response over the course of treatment (6). Suboptimal treatment response can be broadly categorised into two forms; primary resistance and secondary resistance (7), outlined in Table 1.
Full table
Both forms of resistance can be major hurdles for the long-term survival of patients. However, the importance of early treatment response has been increasingly recognised, and typically sets the scene for longer-term outcomes. For example, early molecular response [(EMR) ≤10% BCR-ABL transcript at 3 months] is prognostic of long-term responses, including progression-free and overall survival (8). Unfortunately, one in four patients fail to achieve EMR, and will have poorer long-term prognosis regardless of whether their imatinib dose is increased or are switched to other tyrosine kinase inhibitors (8). Therefore, it is important to optimise treatment early to avoid primary resistance.
Determinants of variable imatinib response
Interpatient variability in imatinib disposition is a major determinant of variable imatinib response, particularly primary resistance. Steady-state trough total plasma imatinib concentrations (Css) can vary more than 25-fold in CML patients administered the same dose (9,10), and are correlated with both CML and GIST treatment responses (5,11). As imatinib has an intracellular site of action, variability in its distribution into CML cells is also a likely, though far less well characterised, contributor to variable response. Therefore, understanding key contributors to interpatient variability in imatinib disposition is essential for the optimisation of its treatment through dose individualisation.
Pharmacodynamic factors, such as acquired kinase domain mutations, play an important role in secondary resistance (7). However, since secondary resistance is unlikely to be overcome by imatinib dose adjustment, and typically necessitates switching to second-line TKIs, these factors are not covered further in this review.
Imatinib pharmacokinetics
Imatinib has high oral bioavailability (95–100%), with peak plasma concentrations approximately 2 hours after administration (12,13). It is highly (~95%) bound to plasma proteins, with volume of distribution estimates ranging from ~170–430 L (13). Imatinib total clearance is approximately 9–14 L/h, with a half-life of 12–34 hours, and it is predominantly hepatically (<15% renal) cleared (13,14). With an intrinsic hepatic clearance of approximately 15 L/h (15), imatinib is a low hepatic extraction drug, and therefore steady-state total plasma imatinib concentrations are determined by variability in plasma protein binding and intrinsic hepatic clearance (metabolism and transport).
Importantly, imatinib has an intracellular site of action, and so variability in imatinib distribution into target CML cells is also likely to be relevant for imatinib efficacy. However, at present no in vivo CML cell intracellular concentration-response relationship has been tested or shown empirically.
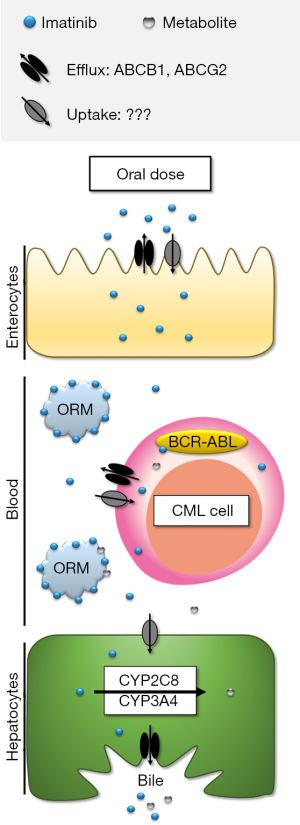
The key contributors to imatinib disposition are thus summarised in Figure 1, and discussed in more detail in the following sections.
Plasma protein binding
Imatinib is highly (~95%) bound to plasma proteins, primarily alpha-1-acid glycoprotein (AAG) (13). Plasma AAG concentrations vary substantially (over 5-fold) between CML patients (16), and will determine the unbound fraction of imatinib available for total clearance and distribution into CML cells. Consequently, variable plasma AAG concentrations contribute significantly to inter-individual variability in total imatinib clearance (~10–20% of co-efficient of variation) (17,18), as well as confounding the total plasma imatinib concentration-response relationship.
Metabolism
Imatinib undergoes hepatic N-demethylation to the much less potent (3- to >10-fold higher IC50) (19-22) major metabolite N-desmethyl imatinib (NDIM); steady-state total plasma NDIM concentrations are approximately 20% that of imatinib (10,23,24). Both imatinib and NDIM undergo mostly hepatic excretion with very little renal contribution (13). Therefore, imatinib biotransformation to NDIM is a clinically important inactivating process, with variable imatinib metabolism likely to be a major contributor to the large inter-patient variability in plasma concentrations (25,26).
In vitro studies using recombinant enzymes indicate that imatinib is N-demethylated by both CYP3A4 and CYP2C8, and possibly CYP3A5, whilst other enzymes (CYP1A2, CYP2D6, CYP2C9 and CYP2C19) play little or no role (27-30). CYP3A4 inhibition studies employing single imatinib doses in healthy participants also indicate a role for CYP3A4 in imatinib in vivo metabolism (31,32). Based on these studies, and the relative abundance of hepatic CYP3A4 compared to CYP2C8, it has long been accepted that CYP3A4 is the major or even sole enzyme responsible for imatinib metabolism in CML patients.
However, steady-state imatinib pharmacokinetics are not significantly influenced by CYP3A4 inducers or inhibitors (13,33,34), and are unrelated to variability in markers of CYP3A activity in CML (35) or GIST patients (36,37). We have also recently demonstrated that imatinib N-demethylation in human liver microsomes is mainly mediated by CYP2C8, and not CYP3A4 (38), potentially as a result of imatinib dose- and time-dependent mechanism-based CYP3A4 inhibition identified in other in vitro studies (29). Consequently, the dominant role of CYP3A4 in imatinib metabolism clinically is coming under question, with CYP2C8 metabolism emerging as a potential major contributor.
Transport
As shown in Figure 1, drug transporters that influence imatinib uptake and efflux could theoretically affect imatinib disposition at multiple levels. For example, uptake transporters in enterocytes could facilitate imatinib absorption. Imatinib is a quadrivalent base [acid dissociation constants (pKa) =1.52, 2.56, 3.73 and 8.07], and is predominantly cationic at pH 6 and below (39). Therefore, it is speculated that an active intestinal uptake process is required to explain imatinib’s high bioavailability. Efflux transporters could conversely limit imatinib absorption, however high imatinib bioavailability would suggest efflux transporters don’t play a significant role in absorption. Drug transporters could also play a role in imatinib uptake and retention in target cancer cells, which has been the focus of extensive in vitro research to date with respect to mechanisms of primary and secondary imatinib resistance (40). Imatinib is partially charged (~33% monocationic) with a distribution co-efficient (logD) of 0.8 at pH 7.4 (41). However, imatinib has a high intracellular: plasma concentration ratio (~8) (42) in patients’ peripheral blood mononuclear cells, indicating an active uptake mechanism. Finally, both influx and efflux transporters expressed on hepatocytes could act to facilitate imatinib biotransformation and excretion, and thus contribute to imatinib clearance.
Evidence for the impact of drug transporters on TKI disposition was recently extensively reviewed (40), and the expert conclusions of Neul and colleagues regarding imatinib can be summarised as follows:
Efflux: imatinib is a substrate of the ABCB1 (P-glycoprotein) and ABCG2 (Breast Cancer Resistance Protein, BCRP) efflux transporters in vitro, and its distribution is significantly altered in Abcb1 and/or Abcg2 knockout mice.
Uptake: whilst SLC22A1 (organic cation transporter, OCT1) has long been touted as a key imatinib transporter, the majority of in vitro and in vivo evidence now indicates that OCT1 is not a significant contributor to imatinib uptake. Similarly, no other uptake transporters investigated to date (SLCO1A2, SLCO1B1, SLCO1B3, SLC22A2-8, SLC47A1) significantly influence imatinib intracellular accumulation. Thus the major transporter(s) responsible for imatinib uptake remains unknown.
We support Neul and colleagues’ recommendations for more appropriate, well-designed, controlled and standardised transporter assays that properly characterise the transport of imatinib. As detailed below, much time and resources may have been misdirected on pharmacogenetic studies of transporters now considered irrelevant to imatinib disposition. No studies have to date investigated the influence of transporter variability (e.g., expression, inhibition, genetics) on imatinib intracellular concentrations in patient cells. This, alongside demonstrating an imatinib intracellular concentration-response relationship clinically, will be critical for translating the extensive in vitro and pharmacogenetic research on transporter-mediated mechanisms of imatinib resistance into improvements in patient outcomes.
Pharmacogenetic studies
Patient germline genetics can potentially play a role in primary resistance. Polymorphisms in genes involved in plasma protein binding (ORM1), metabolism (CYPs) and transport (e.g., ABCB1, ABCG2) are hypothesised to influence the relationship between imatinib dose and total plasma imatinib concentrations. Polymorphisms in genes of transporters expressed in CML cells are also hypothesised to influence intracellular distribution, and thus the plasma concentration-response relationship. Each of these key factors will be discussed in turn.
Plasma protein binding genetics
Plasma AAG comprises a combination of ORM1 and ORM2 gene products (orosomucoid-1 and -2, respectively); imatinib binds primarily to orosomucoid-1 (ORM1) (43). Concentrations and ratios of ORM1 and orosomucoid-2 vary between individuals, and are altered significantly by diseases such as cancer (43,44). In addition, there are three ORM1 haplotypes determined by non-synonymous polymorphisms at two loci (rs17650 113G>A Arg38Gln and rs1126801 Val174Met); *F1 (38Gln-174Val), *F2 (38Gln-174Met) and *S (38Arg-174Val). The 38Gln allele is common (50–70%) and has been reported to influence the unbound fraction of quinidine (45) and pharmacokinetics of telmisartan (46), whilst the 174Met allele is rare (0–5% globally). Thus, in addition to variability in AAG expression, polymorphisms in the ORM1 gene could further increase inter-individual variability in imatinib unbound fraction, and thus total imatinib clearance, further confounding the total plasma imatinib concentration-response relationship. To date however, the impact of ORM1 polymorphisms on imatinib unbound fraction have yet to be characterised in vitro. In the only clinical study to date, Petain and colleagues [2008] found no significant difference in population-pharmacokinetic model predictions of imatinib clearance between ORM1 genotypes among a small sample of 31 paediatric (n=15 *F1/*F1, 1 *F1/*F2, 7 *F1/*S, 7 *S/*S) and 15 adult (n=8 *F1/*F1, 1 *F1/*F2, 3 *F1/*S, 3 *S/*S) GIST patients; imatinib clearance was however significantly negatively correlated with plasma AAG concentrations (17).
Metabolism genetics
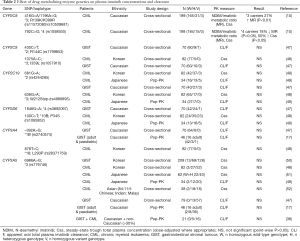
Pharmacogenetic studies investigating CYP450 enzyme polymorphisms are summarised in Table 2.
Full table
CYP3A4/5
Consistent with no impact of CYP3A inhibitors or inducers on imatinib steady-state pharmacokinetics, CYP3A genetics consistently has no significant effect on imatinib pharmacokinetics in CML or GIST patients (Table 2). The major polymorphism influencing variability in CYP3A metabolism is CYP3A5*3 (rs776746, 6986A>G). CYP3A5*3 homozygotes lack functional CYP3A5, whilst heterozygous and wild-type individuals produce relatively high levels of functional CYP3A5. Despite this, CYP3A5*3 genotypes have no significant effect on imatinib pharmacokinetics regardless of the population studied (Caucasian or Asian, CML or GIST).
CYP3A4 variability is poorly defined by common genetic polymorphisms, with CYP3A4 having a relatively low frequency of reduced function allelic variants (53). The two CYP3A4 polymorphisms (*1B and *18) investigated to date have uncertain functional consequences generally, and had no significant effect on imatinib pharmacokinetics. The more recently identified CYP3A4*22 polymorphism, associated with decreased CYP3A4 activity and alterations in substrate pharmacokinetics (53), has yet to be investigated with respect to imatinib. However, any potential clinically relevant effect on imatinib pharmacokinetics is dependent on whether CYP3A4 metabolism actually plays a major role in steady-state imatinib clearance.
CYP2C8
CYP2C8*3 and *4 are the major CYP2C8 polymorphisms in Caucasians. CYP2C8*3 is associated with increased or decreased metabolism in vitro depending on the substrate, and significant effects on pharmacokinetics (54-62). We initially demonstrated that CYP2C8*3 is a gain-of-function haplotype for imatinib N-demethylation in vitro, and have subsequently shown this to translate into significantly increased imatinib metabolism clinically (10,38).
Conversely, CYP2C8*4 is typically associated with decreased activity (55,63-65), and we have shown that CML patients carrying the *4 allele have significantly decreased imatinib metabolism (10). CYP2C8*4 carriers also had 50% higher total plasma imatinib concentrations and were significantly more likely to achieve a study target concentration of 1,000 ng/mL, with all carriers reaching this threshold associated with improved long-term treatment outcomes (10).
Therefore, CYP2C8 genotyping could foreseeably inform imatinib personalised dosing if these findings are replicated.
Other CYP enzymes
Despite playing little or no role in imatinib metabolism, CYP2C9, CYP2C19 and CYP2D6 genotype effects on imatinib pharmacokinetics have also been investigated (47-49), but as expected, no significant associations were identified (Table 2).
Transporter genetics
More than 30 clinical studies have been published investigating whether drug transporter genetic variability influences imatinib disposition. These have either studied genotype differences in the dose-plasma imatinib concentration relationship directly, or intracellular imatinib concentrations tenuously via genotype associations with treatment response. Each of these aspects of imatinib transporter pharmacogenetics is therefore discussed separately.
Effect of transporter genetics on plasma imatinib concentrations and clearance
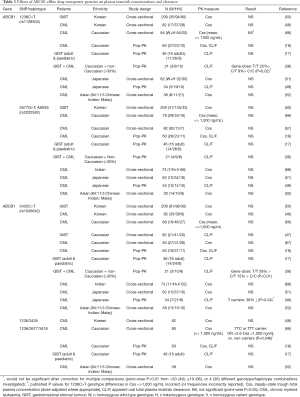
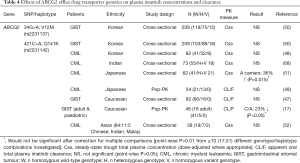
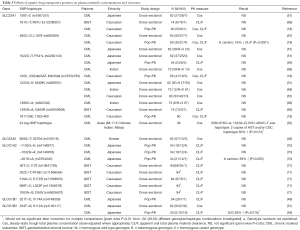
Pharmacogenetic studies investigating the effect of drug transporter gene polymorphisms on plasma imatinib concentration and clearance are summarised in Tables 3-5.
Full table
Full table
Full table
Three major polymorphisms of ABCB1 (1236C>T, 2677G>T and 3435C>T) have been investigated with respect to imatinib disposition, either individually or as a haplotype (Table 3). The expected functional consequences of these polymorphisms for imatinib are not necessarily clear from in vitro and clinical studies of other substrates, although variant ABCB1 genotypes and haplotypes at these loci are typically expected to result in decreased transporter expression (3435C>T) and/or decreased function (1236C>T, 2677G>T, 3435C>T) (72,73). They have therefore been hypothesised to reduce imatinib clearance. However, nearly all studies have found no significant effect of ABCB1 genotype or haplotype on plasma imatinib concentrations or clearance (Table 3).
In a small (n=21) mixed ethnicity and combined CML and GIST patient population, Gurney and colleagues [2007] reported reduced imatinib steady-state clearance estimates for variant ABCB1 1236C>T and 3435C>T genotypes. The proposed mechanism was a reduced imatinib clearance from first dose to steady-state in ABCB1 wild-type, but not variant, patients. However, the 1236C>T association would not be significant after a Bonferroni adjustment for multiple testing [4 ABCB1 and CYP3A5 polymorphisms were investigated in the study (36)], and these positive findings have not been replicated in multiple larger studies. Only two other studies have reported a significant ABCB1 genotype or haplotype effect, and these have been contradictory to the findings of Gurney et al. [2007]. The ABCB1 3435C>T variant allele has been linked to reduced imatinib clearance in Japanese CML patients (49), whilst variant ABCB1 haplotypes have been linked to increased likelihood of plasma imatinib concentrations greater than 1,000 ng/mL in Caucasian CML patients (66). Again, neither of these findings would be significant if adjustments were made for multiple comparisons within the respective studies (Table 3).
Results have been similar for ABCG2, with predominantly negative findings, particularly when accounting for multiple testing within studies (Table 4).
The best characterised polymorphisms in ABCG2 are the non-synonymous 421C>A (Q141K) and 34G>A (V12M), both of which have been investigated for their effect on imatinib disposition. The ABCG2 421C>A polymorphism affects the ATP-binding site of the transporter leading to altered transport of some substrates (74), however no genotype differences in plasma imatinib concentrations or clearance have been found among Korean, Indian or Chinese patients (Table 4). Petain et al. [2008] reported reduced imatinib clearance for the 421 C/A genotype among a combined adult and paediatric Caucasian GIST patient population; however this would not have been statistically significant after adjustment for multiple testing (17). An earlier larger study in Caucasian GIST patients by Gardner et al. [2006] also found no significant ABCG2 421C>A genotype difference in imatinib clearance (47). Findings in Japanese patients have been mixed, with one study reporting increased plasma imatinib concentrations in ABCG2 421A carriers (51), and one reporting no genotype difference in imatinib clearance, albeit with a smaller sample size (n=34) (49).
The ABCG2 34G>A polymorphism causes an amino acid change in the N-terminal intracellular region of the transporter, although the functional consequences of this change appear to be minor (74). Reflecting this, no significant ABCG2 34G>A genotype differences in plasma imatinib concentrations were observed among 209 Korean GIST patients (50).
Aside from the well-studied ABCB1 and ABCG2 efflux transporters, a single cross-sectional study in 62 Japanese CML patients found no significant ABCC2 (MRP2 efflux transporter) 24C>T (rs717620) genotype difference in plasma imatinib concentrations (51). There have also been predominantly negative findings for the influx (uptake) transporter genes (SLC22A1, SLC22A2, SLCO1A2, SLCO1B1 and SLCO1B3) investigated to date (Table 5), reflecting a lack of evidence for their significant role in imatinib transport.
Effect of transporter genetics on imatinib intracellular distribution
No studies have directly investigated whether transporter genetics influence imatinib concentrations within patients’ CML cells. Whilst Nambu and colleagues [2011] investigated associations between SLC22A1, SLCO1B1, SLCO1B3, ABCB1 and ABCG2 polymorphisms and leukocyte intracellular imatinib concentrations in CML patients, patient cells were isolated 3 to 84 (median 19) months into treatment when CML cells make up only a small fraction of circulating cells, and there were no significant genotype differences after accounting for multiple testing (point-wise P≥0.02 from six different polymorphisms and multiple endpoints) (75).
Rather than investigating intracellular distribution directly, previous studies have instead assessed whether transporter genotypes differ in various measures of treatment response. Findings of individual studies investigating the ABCB1 1236C>T, 2677G>T and 3435C>T polymorphisms have been inconsistent and often contradictory, whilst ABCG2 421C>A variant genotypes have generally been associated with either improved or no difference in treatment response (40). Reflecting this, meta-analyses suggest that the ABCG2 421C>A, and less so ABCB1, polymorphisms correlate with imatinib response, at least in Asian CML patients (76-78). Where polymorphisms in uptake transporter genes (SLC22A1, SLCO1A2, SLCO1B3) have been investigated in multiple studies, the clear majority of findings are negative (40). An exception is the SLC22A4 1507C>T polymorphism, which has been associated with poorer treatment response [reduced likelihood of major molecular response (79), and shorter time to disease progression (80)] in two separate studies of Caucasian CML patients.
Unfortunately, despite conclusions often drawn from reported transporter genotype-response relationships, study design limitations have meant that they do not provide strong evidence for a genetic mechanism of variable imatinib intracellular distribution. This is because almost none have controlled for potential genotype effects on plasma imatinib concentrations [with the exception of Vine et al. 2014 (67)], or quantified intracellular imatinib concentrations. Therefore, where genotype correlations with treatment response are identified, it is unknown if this is due to genotype effects on the dose-plasma concentration relationship, genotype effects on imatinib intracellular distribution, or possibly neither (e.g., due to spurious associations, or potential genotype effects on CML pathology unrelated to imatinib disposition, particularly with respect to transporters lacking good evidence for imatinib transport). Without demonstrating a feasible underlying mechanism(s), and hence whether dose adjustment might be of benefit, it is not reasonable to make personalised dosing recommendations based on these associations alone.
Summary of pharmacogenetic studies
CYP2C8 genotype was recently found to significantly affect imatinib metabolism and consequently imatinib exposure in CML patients; a novel finding awaiting replication. Alternatively, multiple studies clearly demonstrate that the CYP3A5*3 polymorphism has no significant effect on imatinib pharmacokinetics clinically. The imatinib efflux transporters ABCB1 and ABCG2 have been well represented in imatinib pharmacogenetic research, whilst many studies have also been devoted to genes encoding transporters with little evidence for, or with evidence against, imatinib transport. Regardless, transporter genetic variability appears to have no reproducible effect on plasma imatinib concentrations or clearance. Whilst the ABCG2 421C>A variant appears to be associated with improved treatment outcomes in Asian CML patients, the mechanism of this association is unknown, and study design limitations have meant that very little is known about whether transporter genetic variability affects imatinib distribution into patients’ CML cells in vivo.
Future directions
A potential caveat to concluding that CYP3A5 and transporter genetics do not influence plasma imatinib concentrations or clearance is that nearly all studies [bar (50)] have been conducted with relatively small sample sizes [median n=68 (range, 21–94)], and all without exception have measured total plasma concentrations likely to be confounded by variability in plasma protein binding. Therefore, these studies have generally lacked sufficient statistical power. In addition to the potential application of meta-analyses to existing imatinib pharmacogenetic data, it is important that future prospective studies are sufficiently powered, particularly taking into account statistical multiple testing.
In order to establish whether genetics influence imatinib intracellular distribution and thus the plasma concentration-response relationship, it should first be demonstrated that an in vivo CML cell intracellular concentration-response relationship exists; at present this relationship is entirely, though soundly, theoretical. Subsequent (or parallel) pharmacogenetic studies will also need to be better designed to include measurements of total plasma imatinib concentrations at a minimum, but ideally unbound plasma and intracellular imatinib concentrations, alongside measures of clinical and molecular response. Our knowledge of genetic risk factors for imatinib adverse effects is also currently limited, with few pharmacogenetics studies having incorporated measures of imatinib toxicity or related dose reduction (10,36,68,81), and no significant findings to date.
In addition to replicating findings for CYP2C8, other novel candidate genes may also warrant investigation. For example, the POR gene encodes the cytochrome p450 oxidoreductase (POR) that provides electrons for microsomal cytochrome P450 metabolism, and the common POR*28 variant has a significant effect on POR activity (82-84). Polymorphisms in genes encoding the nuclear receptors that regulate expression of drug metabolizing enzymes and transporters (e.g., NR1I2, NR1I3, NR1H4, NR3C1, HNF4A, VDR, PPARG) may also be important, and have previously been linked to altered pharmacokinetics of other drugs (85). Although, a small cross-sectional study in 38 mixed ethnicity CML patients found no significant effect of NR1I2 polymorphisms on plasma imatinib concentrations (52).
The identification of the major imatinib uptake transporter(s) will also be of significant importance to our understanding of imatinib disposition, and might also be a source of genetic variability contributing to variable imatinib pharmacokinetics and treatment response.
In moving toward developing tools to improve personalised imatinib dosing, it will also be important to consider integrating pharmacogenetic testing and potential therapeutic drug monitoring or target concentration intervention approaches.
Conclusions
In conclusion, CYP2C8 genotype has a potentially clinically relevant effect on the imatinib dose-plasma concentration relationship, and warrants further investigation. Other drug metabolism and transport genes investigated to date have little or no effect on imatinib clearance. The genetic influence on imatinib intracellular distribution is currently unknown due to major study design limitations. Therefore, whilst potential genetic influences on plasma imatinib exposure have been identified, evidence is still lacking to support a role of pharmacogenetics in personalised imatinib dosing.
Acknowledgments
Funding: This work was supported by the Royal Adelaide Hospital Research Foundation (5256 Award No: 1763).
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Michael Sorich and Andrew Rowland) for the series “Precision dosing of targeted anticancer drugs” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.09.08). The series “Precision dosing of targeted anticancer drugs” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Maino E, Sancetta R, Viero P, et al. Current and future management of Ph/BCR-ABL positive ALL. Expert Rev Anticancer Ther 2014;14:723-40. [Crossref] [PubMed]
- Jabbour E, Kantarjian H. Chronic myeloid leukemia: 2014 update on diagnosis, monitoring, and management. Am J Hematol 2014;89:547-56. [Crossref] [PubMed]
- Linch M, Claus J, Benson C. Update on imatinib for gastrointestinal stromal tumors: duration of treatment. Onco Targets Ther 2013;6:1011-23. [PubMed]
- Gleevec NDA 021588 Label Information Revised Sept 2016. US Food and Drug Administration, Center for Drug Evaluation and Research. 2016. Available online: http://www.accessdata.fda.gov/drugsatfda_docs/label/2016/021588s047lbl.pdf
- Gotta V, Bouchet S, Widmer N, et al. Large-scale imatinib dose-concentration-effect study in CML patients under routine care conditions. Leuk Res 2014;38:764-72. [Crossref] [PubMed]
- Baccarani M, Deininger MW, Rosti G, et al. European LeukemiaNet recommendations for the management of chronic myeloid leukemia: 2013. Blood 2013;122:872-84. [Crossref] [PubMed]
- Baccarani M, Castagnetti F, Gugliotta G, et al. Definition and treatment of resistance to tyrosine kinase inhibitors in chronic myeloid leukemia. Expert Rev Hematol 2014;7:397-406. [Crossref] [PubMed]
- Marin D, Ibrahim AR, Lucas C, et al. Assessment of BCR-ABL1 transcript levels at 3 months is the only requirement for predicting outcome for patients with chronic myeloid leukemia treated with tyrosine kinase inhibitors. J Clin Oncol 2012;30:232-8. [Crossref] [PubMed]
- Larson RA, Druker BJ, Guilhot F, et al. Imatinib pharmacokinetics and its correlation with response and safety in chronic-phase chronic myeloid leukemia. Blood 2008;111:4022-8. [Crossref] [PubMed]
- Barratt DT, Cox HK, Menelaou A, et al. CYP2C8 Genotype significantly alters imatinib metabolism in chronic myeloid leukaemia patients. Clin Pharmacokinet 2017;56:977-85. [Crossref] [PubMed]
- Widmer N, Bardin C, Chatelut E, et al. Review of therapeutic drug monitoring of anticancer drugs part two--targeted therapies. Eur J Cancer 2014;50:2020-36. [Crossref] [PubMed]
- Peng B, Dutreix C, Mehring G, et al. Absolute bioavailability of imatinib (Glivec) orally versus intravenous infusion. J Clin Pharmacol 2004;44:158-62. [Crossref] [PubMed]
- Di Gion P, Kanefendt F, Lindauer A, et al. Clinical pharmacokinetics of tyrosine kinase inhibitors: focus on pyrimidines, pyridines and pyrroles. Clin Pharmacokinet 2011;50:551-603. [Crossref] [PubMed]
- Peng B, Hayes M, Resta D, et al. Pharmacokinetics and pharmacodynamics of imatinib in a phase I trial with chronic myeloid leukemia patients. J Clin Oncol 2004;22:935-42. [Crossref] [PubMed]
- Filppula A. Role of CYP2C8 in the metabolism of montelukast and imatinib, studies in vitro, in silico and in humans. Helsinki: University of Helsinki; 2014.
- Gandia P, Arellano C, Lafont T, et al. Should therapeutic drug monitoring of the unbound fraction of imatinib and its main active metabolite N-desmethyl-imatinib be developed? Cancer Chemother Pharmacol 2013;71:531-6. [Crossref] [PubMed]
- Petain A, Kattygnarath D, Azard J, et al. Population pharmacokinetics and pharmacogenetics of imatinib in children and adults. Clin Cancer Res 2008;14:7102-9. [Crossref] [PubMed]
- Di Paolo A, Polillo M, Capecchi M, et al. The c.480C>G polymorphism of hOCT1 influences imatinib clearance in patients affected by chronic myeloid leukemia. Pharmacogenomics J 2014;14:328-35. [Crossref] [PubMed]
- Mlejnek P, Dolezel P, Faber E, et al. Interactions of N-desmethyl imatinib, an active metabolite of imatinib, with P-glycoprotein in human leukemia cells. Ann Hematol 2011;90:837-42. [Crossref] [PubMed]
- Manley PW, Blasco F, Mestan J, et al. The kinetic deuterium isotope effect as applied to metabolic deactivation of imatinib to the des-methyl metabolite, CGP74588. Bioorg Med Chem 2013;21:3231-9. [Crossref] [PubMed]
- Skoglund K, Boiso Moreno S, Jonsson JI, et al. Single-nucleotide polymorphisms of ABCG2 increase the efficacy of tyrosine kinase inhibitors in the K562 chronic myeloid leukemia cell line. Pharmacogenet Genomics 2014;24:52-61. [Crossref] [PubMed]
- Skoglund K, Moreno SB, Baytar M, et al. ABCB1 haplotypes do not influence transport or efficacy of tyrosine kinase inhibitors in vitro. Pharmgenomics Pers Med 2013;6:63-72. [Crossref] [PubMed]
- Josephs DH, Fisher DS, Spicer J, et al. Clinical pharmacokinetics of tyrosine kinase inhibitors: implications for therapeutic drug monitoring. Ther Drug Monit 2013;35:562-87. [PubMed]
- Gschwind HP, Pfaar U, Waldmeier F, et al. Metabolism and disposition of imatinib mesylate in healthy volunteers. Drug Metab Dispos 2005;33:1503-12. [Crossref] [PubMed]
- Bouchet S, Titier K, Moore N, et al. Therapeutic drug monitoring of imatinib in chronic myeloid leukemia: experience from 1216 patients at a centralized laboratory. Fundam Clin Pharmacol 2013;27:690-7. [Crossref] [PubMed]
- de Wit D, Guchelaar HJ, den Hartigh J, et al. Individualized dosing of tyrosine kinase inhibitors: are we there yet? Drug Discov Today 2015;20:18-36. [Crossref] [PubMed]
- Filppula AM, Neuvonen M, Laitila J, et al. Autoinhibition of CYP3A4 Leads to Important Role of CYP2C8 in Imatinib Metabolism. Drug Metab Dispos 2013;41:50-9. [Crossref] [PubMed]
- Rochat B, Zoete V, Grosdidier A, et al. In vitro biotransformation of imatinib by the tumor expressed CYP1A1 and CYP1B1. Biopharm Drug Dispos 2008;29:103-18. [Crossref] [PubMed]
- Filppula AM, Laitila J, Neuvonen PJ, et al. Potent mechanism-based inhibition of CYP3A4 by imatinib explains its liability to interact with CYP3A4 substrates. Br J Pharmacol 2012;165:2787-98. [Crossref] [PubMed]
- Nebot N, Crettol S, d'Esposito F, et al. Participation of CYP2C8 and CYP3A4 in the N-demethylation of imatinib in human hepatic microsomes. Br J Pharmacol 2010;161:1059-69. [Crossref] [PubMed]
- Bolton AE, Peng B, Hubert M, et al. Effect of rifampicin on the pharmacokinetics of imatinib mesylate (Gleevec, STI571) in healthy subjects. Cancer Chemother Pharmacol 2004;53:102-6. [Crossref] [PubMed]
- Dutreix C, Peng B, Mehring G, et al. Pharmacokinetic interaction between ketoconazole and imatinib mesylate (Glivec) in healthy subjects. Cancer Chemother Pharmacol 2004;54:290-4. [Crossref] [PubMed]
- Schmidli H, Peng B, Riviere GJ, et al. Population pharmacokinetics of imatinib mesylate in patients with chronic-phase chronic myeloid leukaemia: results of a phase III study. Br J Clin Pharmacol 2005;60:35-44. [Crossref] [PubMed]
- van Erp NP, Gelderblom H, Karlsson MO, et al. Influence of CYP3A4 inhibition on the steady-state pharmacokinetics of imatinib. Clin Cancer Res 2007;13:7394-400. [Crossref] [PubMed]
- Skoglund K, Richter J, Olsson-Stromberg U, et al. In vivo cytochrome P450 3A isoenzyme activity and pharmacokinetics of imatinib in relation to therapeutic outcome in patients with chronic myeloid leukemia. Ther Drug Monit 2016;38:230-8. [Crossref] [PubMed]
- Gurney H, Wong M, Balleine RL, et al. Imatinib disposition and ABCB1 (MDR1, P-glycoprotein) genotype. Clin Pharmacol Ther 2007;82:33-40. [Crossref] [PubMed]
- Widmer N, Decosterd LA, Csajka C, et al. Population pharmacokinetics of imatinib and the role of alpha-acid glycoprotein Br J Clin Pharmacol 2006;62:97-112. [published erratum appears in Br J Clin Pharmacol 2010 Aug; 70 (2): 316]. [Crossref] [PubMed]
- Khan MS, Barratt DT, Somogyi AA. Impact of CYP2C8*3 polymorphism on in vitro metabolism of imatinib to N-desmethyl imatinib. Xenobiotica 2016;46:278-87. [Crossref] [PubMed]
- Szakács Z, Béni S, Varga Z, et al. Acid-base profiling of imatinib (gleevec) and its fragments. J Med Chem 2005;48:249-55. [Crossref] [PubMed]
- Neul C, Schaeffeler E, Sparreboom A, et al. Impact of Membrane Drug Transporters on Resistance to Small-Molecule Tyrosine Kinase Inhibitors. Trends Pharmacol Sci 2016;37:904-32. [Crossref] [PubMed]
- Giannoudis A, Davies A, Lucas CM, et al. Effective dasatinib uptake may occur without human organic cation transporter 1 (hOCT1): implications for the treatment of imatinib-resistant chronic myeloid leukemia. Blood 2008;112:3348-54. [Crossref] [PubMed]
- D'Avolio A, Simiele M, De Francia S, et al. HPLC-MS method for the simultaneous quantification of the antileukemia drugs imatinib, dasatinib and nilotinib in human peripheral blood mononuclear cell (PBMC). J Pharm Biomed Anal 2012;59:109-16. [Crossref] [PubMed]
- Fitos I, Visy J, Zsila F, et al. Selective binding of imatinib to the genetic variants of human alpha1-acid glycoprotein. Biochim Biophys Acta 2006;1760:1704-12. [Crossref] [PubMed]
- Hanada K, Yamanaka E, Yamamoto N, et al. Effects of surgery & chronic disease states on the concentrations & phenotype distribution of alpha1-acid glycoprotein: studies in patients with breast cancer & patients with chronic inflammatory disease. Int J Clin Pharmacol Ther 2011;49:415-21. [Crossref] [PubMed]
- Li JH, Xu JQ, Cao XM, et al. Influence of the ORM1 phenotypes on serum unbound concentration and protein binding of quinidine. Clin Chim Acta 2002;317:85-92. [Crossref] [PubMed]
- Chen WQ, Shu Y, Li Q, et al. Polymorphism of ORM1 is associated with the pharmacokinetics of telmisartan. PloS one 2013;8:e70341 [Crossref] [PubMed]
- Gardner ER, Burger H, van Schaik RH, et al. Association of enzyme and transporter genotypes with the pharmacokinetics of imatinib. Clin Pharmacol Ther 2006;80:192-201. [Crossref] [PubMed]
- Seong SJ, Lim M, Sohn SK, et al. Influence of enzyme and transporter polymorphisms on trough imatinib concentration and clinical response in chronic myeloid leukemia patients. Ann Oncol 2013;24:756-60. [Crossref] [PubMed]
- Yamakawa Y, Hamada A, Nakashima R, et al. Association of genetic polymorphisms in the influx transporter SLCO1B3 and the efflux transporter ABCB1 with imatinib pharmacokinetics in patients with chronic myeloid leukemia. Ther Drug Monit 2011;33:244-50. [PubMed]
- Koo DH, Ryu MH, Ryoo BY, et al. Association of ABCG2 polymorphism with clinical efficacy of imatinib in patients with gastrointestinal stromal tumor. Cancer Chemother Pharmacol 2015;75:173-82. [Crossref] [PubMed]
- Takahashi N, Miura M, Scott SA, et al. Influence of CYP3A5 and drug transporter polymorphisms on imatinib trough concentration and clinical response among patients with chronic phase chronic myeloid leukemia. J Hum Genet 2010;55:731-7. [Crossref] [PubMed]
- Singh O, Chan JY, Lin K, et al. SLC22A1-ABCB1 Haplotype Profiles Predict Imatinib Pharmacokinetics in Asian Patients with Chronic Myeloid Leukemia. PloS One 2012;7:e51771 [Crossref] [PubMed]
- Klein K, Zanger UM. Pharmacogenomics of cytochrome P450 3A4: recent progress toward the "missing heritability" problem. Front Genet 2013;4:12. [Crossref] [PubMed]
- Dai D, Zeldin DC, Blaisdell JA, et al. Polymorphisms in human CYP2C8 decrease metabolism of the anticancer drug paclitaxel and arachidonic acid. Pharmacogenetics 2001;11:597-607. [Crossref] [PubMed]
- Gao Y, Liu D, Wang H, et al. Functional characterization of five CYP2C8 variants and prediction of CYP2C8 genotype-dependent effects on in vitro and in vivo drug-drug interactions. Xenobiotica 2010;40:467-75. [Crossref] [PubMed]
- Yu L, Shi D, Ma L, et al. Influence of CYP2C8 polymorphisms on the hydroxylation metabolism of paclitaxel, repaglinide and ibuprofen enantiomers in vitro. Biopharm Drug Dispos 2013;34:278-87. [Crossref] [PubMed]
- Bergmann TK, Brasch-Andersen C, Green H, et al. Impact of CYP2C8*3 on paclitaxel clearance: a population pharmacokinetic and pharmacogenomic study in 93 patients with ovarian cancer. Pharmacogenomics J 2011;11:113-20. [Crossref] [PubMed]
- Gréen H, Söderkvist P, Rosenberg P, et al. Pharmacogenetic studies of Paclitaxel in the treatment of ovarian cancer. Basic Clin Pharmacol Toxicol 2009;104:130-7. [Crossref] [PubMed]
- Daily EB, Aquilante CL. Cytochrome P450 2C8 pharmacogenetics: a review of clinical studies. Pharmacogenomics 2009;10:1489-510. [Crossref] [PubMed]
- Kirchheiner J, Meineke I, Fuhr U, et al. Impact of genetic polymorphisms in CYP2C8 and rosiglitazone intake on the urinary excretion of dihydroxyeicosatrienoic acids. Pharmacogenomics 2008;9:277-88. [Crossref] [PubMed]
- Muschler E, Lal J, Jetter A, et al. The role of human CYP2C8 and CYP2C9 variants in pioglitazone metabolism in vitro. Basic Clin Pharmacol Toxicol 2009;105:374-9. [Crossref] [PubMed]
- Stage TB, Christensen MM, Feddersen S, et al. The role of genetic variants in CYP2C8, LPIN1, PPARGC1A and PPARgamma on the trough steady-state plasma concentrations of rosiglitazone and on glycosylated haemoglobin A1c in type 2 diabetes. Pharmacogenet Genomics 2013;23:219-27. [Crossref] [PubMed]
- Bahadur N, Leathart JB, Mutch E, et al. CYP2C8 polymorphisms in Caucasians and their relationship with paclitaxel 6alpha-hydroxylase activity in human liver microsomes. Biochem Pharmacol 2002;64:1579-89. [Crossref] [PubMed]
- Jiang H, Zhong F, Sun L, et al. Structural and functional insights into polymorphic enzymes of cytochrome P450 2C8. Amino acids 2011;40:1195-204. [Crossref] [PubMed]
- Wang Z, Wang S, Huang M, et al. Characterizing the effect of cytochrome P450 (CYP) 2C8, CYP2C9, and CYP2D6 genetic polymorphisms on stereoselective N-demethylation of fluoxetine. Chirality 2014;26:166-73. [Crossref] [PubMed]
- Dulucq S, Bouchet S, Turcq B, et al. Multidrug resistance gene (MDR1) polymorphisms are associated with major molecular responses to standard-dose imatinib in chronic myeloid leukemia. Blood 2008;112:2024-7. [Crossref] [PubMed]
- Vine J, Cohen SB, Ruchlemer R, et al. Polymorphisms in the human organic cation transporter and the multidrug resistance gene: correlation with imatinib levels and clinical course in patients with chronic myeloid leukemia. Leuk Lymphoma 2014;55:2525-31. [Crossref] [PubMed]
- Francis J, Dubashi B, Sundaram R, et al. A study to explore the correlation of ABCB1, ABCG2, OCT1 genetic polymorphisms and trough level concentration with imatinib mesylate-induced thrombocytopenia in chronic myeloid leukemia patients. Cancer Chemother Pharmacol 2015;76:1185-9. [Crossref] [PubMed]
- Hu S, Franke RM, Filipski KK, et al. Interaction of imatinib with human organic ion carriers. Clin Cancer Res 2008;14:3141-8. [Crossref] [PubMed]
- Yamakawa Y, Hamada A, Shuto T, et al. Pharmacokinetic impact of SLCO1A2 polymorphisms on imatinib disposition in patients with chronic myeloid leukemia. Clin Pharmacol Ther 2011;90:157-63. [Crossref] [PubMed]
- Eechoute K, Franke RM, Loos WJ, et al. Environmental and genetic factors affecting transport of imatinib by OATP1A2. Clin Pharmacol Ther 2011;89:816-20. [Crossref] [PubMed]
- Ieiri I. Functional significance of genetic polymorphisms in P-glycoprotein (MDR1, ABCB1) and breast cancer resistance protein (BCRP, ABCG2). Drug Metab Pharmacokinet 2012;27:85-105. [Crossref] [PubMed]
- Wolf SJ, Bachtiar M, Wang J, et al. An update on ABCB1 pharmacogenetics: insights from a 3D model into the location and evolutionary conservation of residues corresponding to SNPs associated with drug pharmacokinetics. Pharmacogenomics J 2011;11:315-25. [Crossref] [PubMed]
- Bruhn O, Cascorbi I. Polymorphisms of the drug transporters ABCB1, ABCG2, ABCC2 and ABCC3 and their impact on drug bioavailability and clinical relevance. Expert Opin Drug Metab Toxicol 2014;10:1337-54. [Crossref] [PubMed]
- Nambu T, Hamada A, Nakashima R, et al. Association of SLCO1B3 polymorphism with intracellular accumulation of imatinib in leukocytes in patients with chronic myeloid leukemia. Biol Pharm Bull 2011;34:114-9. [Crossref] [PubMed]
- Jiang ZP, Zhao XL, Takahashi N, et al. Trough concentration and ABCG2 polymorphism are better to predict imatinib response in chronic myeloid leukemia: a meta-analysis. Pharmacogenomics 2017;18:35-56. [Crossref] [PubMed]
- Zheng Q, Wu H, Yu Q, et al. ABCB1 polymorphisms predict imatinib response in chronic myeloid leukemia patients: a systematic review and meta-analysis. Pharmacogenomics J 2015;15:127-34. [Crossref] [PubMed]
- Zu B, Li Y, Wang X, et al. MDR1 gene polymorphisms and imatinib response in chronic myeloid leukemia: a meta-analysis. Pharmacogenomics 2014;15:667-77. [Crossref] [PubMed]
- Angelini S, Soverini S, Ravegnini G, et al. Association between imatinib transporters and metabolizing enzymes genotype and response in newly diagnosed chronic myeloid leukemia patients receiving imatinib therapy. Haematologica 2013;98:193-200. [Crossref] [PubMed]
- Angelini S, Pantaleo MA, Ravegnini G, et al. Polymorphisms in OCTN1 and OCTN2 transporters genes are associated with prolonged time to progression in unresectable gastrointestinal stromal tumours treated with imatinib therapy. Pharmacol Res 2013;68:1-6. [Crossref] [PubMed]
- Au A, Aziz Baba A, Goh AS, et al. Association of genotypes and haplotypes of multi-drug transporter genes ABCB1 and ABCG2 with clinical response to imatinib mesylate in chronic myeloid leukemia patients. Biomed Pharmacother 2014;68:343-9. [Crossref] [PubMed]
- Huang N, Agrawal V, Giacomini KM, et al. Genetics of P450 oxidoreductase: sequence variation in 842 individuals of four ethnicities and activities of 15 missense mutations. Proc Natl Acad Sci U S A 2008;105:1733-8. [Crossref] [PubMed]
- Sandee D, Morrissey K, Agrawal V, et al. Effects of genetic variants of human P450 oxidoreductase on catalysis by CYP2D6 in vitro. Pharmacogenet Genomics 2010;20:677-86. [Crossref] [PubMed]
- Oneda B, Crettol S, Jaquenoud Sirot E, et al. The P450 oxidoreductase genotype is associated with CYP3A activity in vivo as measured by the midazolam phenotyping test. Pharmacogenet Genomics 2009;19:877-83. [Crossref] [PubMed]
- Urquhart BL, Tirona RG, Kim RB. Nuclear receptors and the regulation of drug-metabolizing enzymes and drug transporters: implications for interindividual variability in response to drugs. J Clin Pharmacol 2007;47:566-78. [Crossref] [PubMed]


