Methods for quantification and characterization of microRNAs in cell-free plasma/serum, normal exosomes and tumor-derived exosomes
Introduction
The detection of extracellular nucleic acids in human blood circulation dates back nearly 70 years (1). More than 40 years later, the importance of circulating nucleic acids was recognized by the detection of mutated DNA molecules in the bloodstream of cancer patients (2,3). Nowadays, it is documented that under pathological circumstances, increased amounts of nucleic acids are released into the cell microenvironment by various cell physiological events, including apoptosis, necrosis and active secretion (4,5). In particular, cancer patients display high levels of nucleic acids in their blood circulation. These high concentrations in blood may originate from the primary tumor, circulating tumor cells (CTCs) and micrometastatic deposits at distant sites (6,7). Nucleic acids may also be shed into the tumor microenvironment by immunocytes and stromal cells or from other affected organs that mediate cellular responses to tumor burden and inflammatory reactions (8), but a proportion of these molecules may also be derived from healthy organs. Thus, blood of cancer patients may constitute a pool of circulating cancer-derived and wild-type (normal) nucleic acids discharged from different sources.
Not until 2008, the presence of cell-free microRNAs (miRNAs) in blood was described. This was the first evidence showing the feasibility of quantifying circulating miRNAs from serum. In this study, the association of high expression levels of miR-21 with relapse-free survival in patients with diffuse large B-cell lymphoma was reported (9). To date, numerous studies have been published, suggesting that deregulated levels of miRNAs are related to tumor classification, diagnosis, disease progression, and prognosis (10). In blood, miRNAs circulate in a highly stable form and are protected against RNase digestion, presumably because most of them are incorporated in apoptotic bodies, exosomes, and/or complexed with RNA-binding proteins, such as Argonaute 2 (AGO2) and HDL proteins (11).
The discovery of exosomes in the early 1980s was not tracked by great attention, since at that time, exosomes were assumed to be unnecessary cellular components (12). In 2007, a year earlier than the detection of cell-free miRNAs in blood, Valadi et al. introduced the term “exosomal shuttle RNA”. For the first time, the authors demonstrated that exosomes contain miRNAs that can be transported from a donor to a recipient cell, and be functional in the new location (13). It is assumed that miRNAs are selectively packaged into exosomes and that their active secretion in exosomes seems to be a regulated process directly linked to disease pathogenesis (14). As a result, the selective exosomal miRNA transfer causes a decrease in specific miRNAs in the donor cell and a potential manipulation of the recipient cell, promoting tumorigenesis, tumor progression and metastasis. This exosomal miRNA shuttle makes them attractive candidates for therapeutic targets in clinical applications, but prior to the entry in the clinics, robust extraction and standardized detection methods of these biomolecules have to be established. Therefore, the main concern of the present review article is to introduce and discuss different technical platforms applied and developed for miRNA and exosome analyses.
Characteristics of miRNAs
Currently, there are more than 2,000 different miRNAs that can be found in the databank (
Characteristics of exosomes
The major class of extracellular vesicles comprises exosomes ranging from 30–100 nm in size. Within the endosomal pathway of the cell, exosomes are produced by inward budding of the luminal membrane of multivesicular bodies, a late endosomal compartment. Subsequently, the fusion of the multivesicular bodies with the cytoplasma membrane of the donor cell results in the active secretion of exosomes into the extracellular area and blood circulation (20). Subsequently, the exocytotic exosomes deliver their cargo, including proteins, DNA, RNAs and lipids, to the recipient cell by fusing with its cell membrane. Multiple cell types, including dendritic cells, lymphocytes and tumor cells are involved in this cell-to-cell communication (21). In this manner, exosomes may serve as suppliers of cancer-derived and tumor-suppressive genetic information, and consequently, transform their host cell and surveil the tumor formation, respectively (22). This exosome shuttle affects cellular signaling, metabolic functions and homeostasis (23). Amazingly, exosomes are also able to carry out cell-independent biogenesis, since they contain the RISC complex in which pre-miRNAs are synthesized in mature miRNAs. This miRNA biogenesis in exosomes is assumed to contribute to cancer progression in recipient cells (24).
In cancer patients, the excessive secretion of exosomes has been associated with tumor invasiveness and promotion of increased proliferation and migration of tumor cells leading to metastasis (23,25). Furthermore, the relevance of exosomal miRNAs as diagnostic and therapeutic molecular markers in cancer patients has been suggested by numerous studies (26). During tumor progression, the exosome-mediated secretion of miRNAs may be selected as a mechanism to coordinate activation of the metastatic cascade (27). E.g., the transfer of exosomal miR-105 to non-metastatic breast cancer cells may induce metastasis and vascular permeability in the recipient cells (28). The ability of drug-resistant breast cancer cells to transmit their capacity for resistance to receptor chemosensitive cell lines may be due to the exosome shuttle of miR-100, miR-222 and miR-30a (29,30). Conclusively, exosomal miRNAs may induce an oncogenic field effect in adjacent normal cells to participate in cancer development and progression.
Preanalytical variables for the quantification of exosomes and miRNAs
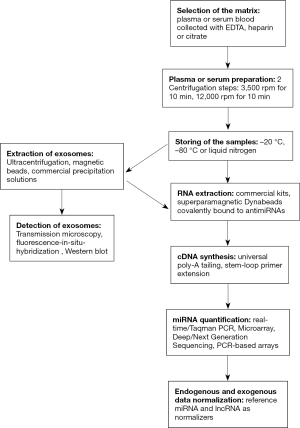
Although nucleic acids are present in many different biological fluids (e.g., saliva, tears, urine, bronchoalveolar lavage, ductal lavage), plasma or serum are most preferred sources for their analysis (6). Prior to the extraction of cell-free and exosomal miRNAs from plasma/serum, the handling and processing of blood specimens should be considered. The time from blood withdrawal to storage, storage temperature, centrifugation speed and centrifugation time are important parameters for plasma/serum sample preparation. For example, the processing time should be preferable less than 2 hours. The first centrifugation step of the blood samples should be at 2,000 g for 10 minutes. The long-term storage temperature should be set at 70–80 °C. After thawing and prior to the use of the samples a second centrifugation step at 2,000 g for 10 minutes should be included. These guidelines should be followed and intensified, since without a global consensus on the procedures the clinical significance of circulating miRNAs may not be proven (31).
Currently, it still remains a matter of debates, whether serum or plasma is the optimal source for the analyses. However, scientists begin to prefer plasma, since it seems to contain less nucleic acids from hemolyzed blood cells that may contaminate and mask tumor-derived nucleic acids. The reason for the lower contamination grade is the preparation of plasma that requires anticoagulants, such as heparin, acid citrate dextrose (ACD) or EDTA. Of note is that a significant release of miRNAs from blood cells was already observed 1 hour after blood collection, and this discharge appears also to contain vesicle-associated miRNAs (32). This incident has, of course, an impact on the pattern of extracellular miRNAs that is highly sensitive to artificial changes caused by blood cell-derived miRNAs after blood puncture. Thus, before starting the experiment, the hemolysis of plasma and serum samples should be examined, to avoid biases in the quantification of circulating miRNAs. This accordingly applies to the analysis of all nucleic acids and proteins.
Hemolysis can simply be assessed by spectrophotometry at wavelengths from 350 to 650 nm. Here, a dilution series of lysed red blood cells serves as a standard curve. The degree of hemolysis is then determined due to the optical density (OD) at 414 nm (absorbance peak of free hemoglobin, called Soret band), along with additional peaks at 541 and 576 nm and the standard curve. Samples are classified as being hemolyzed if the OD at 414 exceeded 0.25 (33). In addition, plasma/serum samples can also be examined for erythrocyte miRNA contamination using the measured values of miR-451 and miR-23a. If the ratio between these values is higher than 5, a possible contamination is indicated (34).
As soon as the preanalytical factors are settled down, the analytical factors which are discussed below have to be considered and standardized.
Quantification of miRNAs
For the extraction of total RNA, several commercial products are offered on the market. They usually enrich RNA by phenol/guanidinium followed by isopropanol precipitation and spin columns, to avoid contamination with genomic DNA. The most commonly used assays are the mirVana kit from Life Technologies and the miRNeasy kit from Qiagen. These kits are particularly qualified for the subsequent quantification of miRNAs using the real-time PCR-based and miRNA-specific assays from the respective company. Another approach is the direct extraction of miRNAs by superparamagnetic Dynabeads covalently bound to two sets of 377 anti-miRNA oligonucleotides from Life Technologies. It relies on hybridization of endogenous miRNAs to the corresponding anti-miRNA oligonucleotides conjugated to these beads. This procedure provides higher miRNA amounts and is more suitable for small input material, since it avoids the circuitous step through the extraction of total RNA. The disadvantage of this technique is the limited number of miRNAs to be quantified. Current methods for conversing RNA/miRNAs into cDNA are universal poly-A tailing and stem-loop primer extension. For amplification, the specificity and sensitivity of stem loop primers are somewhat higher than those of conventional linear primers, since base stacking and spatial constraint of the stem-loop structure improve the thermal stability and prevent it from binding double-strand genomic DNA molecules (35).
For miRNA quantification, there are several different technics depending on the number of miRNAs to be analyzed (36). Quantitative real-time PCR using TaqMan probes or SybrGreen is the gold standard for quantifying miRNAs. These methods generate fluorescence signals that are proportional to the amount of the generated PCR product. Real-time PCR using SybrGreen has a somewhat lower specificity than PCR based on TaqMan probes. The reason of it is that SybrGreen non-specifically binds to DNA, whereas TaqMan probes have a specific DNA binding activity, and rely on fluorescence energy resonance transfer and 5’-nuclease activity of the Taq-polymerase. So far, Northern blot assays that detect both mature and precursor forms of any miRNA have rarely been used. The procedure is more elaborate with its separation of small RNA molecules by gel electrophoresis, their transfer from the gel to a membrane and their hybridization with labeled probes. Disadvantages of this technique are also its low sensitivity and the required input of high amounts of starting material. To profile a large number of different miRNAs, miRNA arrays are frequently used. Microarray-based technologies rely on hybridization of the extracted RNA to specific probes, which can cover more than 1,000 mature human miRNAs sequences listed in the miRNA database (Sanger miRNABase). However, they are challenging to optimize probes and hybridization conditions. Nevertheless, the obtained miRNA array data can be validated by a subsequent quantitative real-time PCR, as described above. Nowadays, the application of microarrays has become more seldom, and has been more and more replaced by real-time PCR-based TaqMan arrays and next-generation sequencing (NGS). Real-time PCR-based TaqMan arrays are much easier in handling, but they are mounted by a smaller number of (e.g., 48, 96 or 384) miRNAs than the classical microarrays. In the last years, NGS, a largely sequence-independent method, has been widely used. The approach comprises the amplification of adapter-ligated sample RNA and cDNA libraries along with a following sequencing step of the PCR products. The output data deliver sequencing reads of varying lengths corresponding to a huge number of miRNAs that allow besides the detection of known miRNAs, the identification of novel miRNAs. Drawbacks of this method are that miRNA sequence biases can be introduced during library construction and that computational support is needed to analyze the extensive data output. Moreover, the high costs of the technical platform and analyses are to take into consideration (36). These challenges have led to the founding of specialized facilities that offer technical and computational services.
Finally, regardless of which technique for miRNA quantification is carried out, a data normalization step with both endogenous and exogenous reference genes should be included, to assess quality and handling of the plasma/serum samples. Endogenous data normalization aims at removing differences due to blood sampling and accounting for quality of the samples, while exogenous normalization accounts for the technical, inter-individual variability. Unfortunately, an appropriate endogenous miRNA control for miRNA data normalization has still not been established, leading to the fact that each laboratory prefers its own endogenous reference gene. However, the wrong choice of a reference gene has a great impact on the study outcome, and is particularly problematic for plasma/sera analyses. Important for a correct data normalization is the algorithms that should dispose stably expressed, endogenous reference genes across all patient and healthy control samples. For exogenous data normalization, synthetic, nonhuman spike-in miRNAs, e.g., the C. elegans miRNA cel-miR-39 can be used to monitor RNA purification and reverse transcription efficiencies. The final, normalized, relative miRNA value refers to the delta Cq (PCR-derived cycle threshold) value which is calculated by the Cq value of the target miRNA minus the average value of the stably expressed endogenous miRNA and the exogenous miRNA (37).
Recovery of exosomes
In addition to human body fluids, among others plasma and serum, miRNAs can also be extracted from exosomes. Diverse exosome extraction techniques are described in the following paragraphs.
Commonly used exosome isolation techniques include ultracentrifugation, density gradient separation, immunoaffinity assays and polymeric methods (38). Verification of the extracted exosomes is usually carried out by microscopy, Western blot or flow cytometry (Figure 1). Among the extraction methods, ultracentrifugation is the most commonly used technology for exosome concentration. This method requires differential centrifugation steps with ultrahigh speeds up to 200,000 g. The performance of this procedure requires an expensive ultracentrifuge and is time consuming taking more than 10 hours. The purity of the ultracentrifuged exosomes can, then, be improved by adding a subsequent sucrose density gradient centrifugation step. Exosomes can also be prepared by filtering them through a porous membrane and subsequently centrifuging them at high speed on a sucrose cushion. Applying a filter with pores in size of 100 nm and two ultracentrifugation steps, Grigor’eva et al. (39) could even isolate exosomes from human tears. Versatile tools are magnetic beads coated with antibodies that capture exosomes by exosomal surface markers (40). Typically exosomal markers are the most abundant membrane proteins of the tetraspanin superfamily. Among them, CD9, CD63, CD81, CD82 and CD151 have a broad tissue distribution, while Tssc6, CD37, and CD53 are restricted to particular tissues, such as hematopoietic cells (41). Heat shock proteins (Hsp60, Hsp70, Hsp90) are also found on exosomes. The composition of all these membrane proteins differs according to their cell or tissue of origin. Most widely used target molecules are CD9, CD63 and CD81 for the extraction of total exosomes independent of their tissue origin.
Kalra et al. (42) performed a comparative evaluation of the three exosome isolation techniques: differential centrifugation coupled with ultracentrifugation, epithelial cell adhesion molecule immunoaffinity pull-down and OptiPrep(TM) density gradient separation using plasma from healthy individuals. Due to Western blotting and microscopy results, they found that the OptiPrep(TM) density gradient method was superior in isolating pure exosomal populations over the other methods. Moreover, the extracted exosomes were not contaminated with the highly abundant plasma proteins. The researchers also assessed the stability of exosomes in plasma under various storage conditions. Western blotting analysis using the exosomal marker TSG101 revealed that exosomes are stable for 90 days.
Among the commercial exosome precipitation kits, the most commonly used kit is the ExoQuick precipitation solution from System Biosciences, a quick but relatively expensive method. ExoQuick implicates a commercial agglutinating agent to precipitate exosomes. Taylor et al. (43) compared the exosome extraction by ExoQuick with that by ultracentrifugation chromatography and magnetic beads. The highest amounts of exosomes could be isolated using ExoQuick than applying the other methods. Moreover, serum delivered higher yields than plasma, possibly because the exosome pellet is easier to dissolve in fibrinogen-free serum than in plasma (Figure 1).
Optical and non-optical methods are carried out, to characterize the extracted exosomes. Transmission electron microscopy certainly displays the most detailed exosome images, and it is applied to determine shape, size and purity of exosomes. Conventional optical methods, such as dynamic light scattering (DLS) and fluorescence microscopy, have indeed lower detection limits. DLS is one of the most commonly used techniques to estimate the size of small particles and the molecular weights of large protein complexes. The fluorescence microscopy that relies on fluorescence-in-situ-hybridization (FISH) is also an easy and quick method to visualize exosomes. Fluorescence dyes, such as Cy3 (red) and FITC (green) are used to label exosomes. Western blotting belongs to the non-optical methods. This commonly used technique is most frequently applied in connection with antibodies against the exosomal markers CD9, CD63, CD81.
Quantification of subpopulations of exosomes
Exosomes are released from all cell types, such as normal cells, cancer cells and stem cells (26,44). Their number and cargo vary depending on cell type and state of health. Thus, exosomes are loaded with biomolecules that may predict the cells of their origin. As exosomes are capable of transferring the malignant phenotype of their donor cells to normal cells, establishing a local and distant microenvironment, and promoting cancer cell growth, tumor progression and metastasis, their profiling may be a promising tool for cancer diagnosis and monitoring of therapeutic efficacy (21). In particular, miRNAs from tumor-derived exosomes may be involved in the neoplastic transformation process of a normal to a malignant cell. Therefore, it is of interest to analyze the expression profiles of miRNAs in the subpopulation of cancer-derived exosomes that are assumed to reflect the characteristics of the primary tumor, CTCs and/or distant metastases (14,25). However, marker proteins that allow enrichment of tumor-derived exosomes over normal exosomes are poorly defined. A potential marker protein could be the epithelial surface antigen (glycoprotein) EpCAM, since it is usually overexpressed in tumors. EpCAM is located in the epithelial intercellular junctions mediating homophilic calcium-dependent cell-cell adhesion (45). It is an important marker for the detection of CTCs that are released by epithelial tumors into the blood circulation and that are probably the origin of intractable metastatic disease (46).
In 2008, Taylor et al. (47) isolated EpCAM-positive exosomes from serum of ovarian cancer patients for the first time, applying a modified magnetic activated cell sorting (MACS) procedure. The levels of 8 miRNAs (miR-21, miR-141, miR-200a, miR-200c, miR-200b, miR-203, miR-205 and miR-214) in EpCAM-positive exosomes were similar to those in primary tumor cell cultures established from ovarian cancer patients. While EpCAM-positive exosomes were detectable in both patients with benign ovarian disease and ovarian cancer, the similar exosomal miRNAs profiles in ovarian cancer patients were significantly distinct from those observed in benign disease. In age-matched, healthy volunteers, the levels of EpCAM-positive exosomes corresponded to the background of the assay. These findings demonstrate that EpCAM-positive exosomes may predominantly be tumor-derived. In 2011, Rupp et al. (48) characterized CD24, a small but extensively glycosylated protein linked to the cell surface by a glycosylphosphatidylinositol anchor, as a more useful marker than EpCAM for the extraction of tumor-derived exosomes using Western blot analysis and antibody coupled magnetic beads. They observed that in breast cancer patients CD24 was present but EpCAM was absent on serum exosomes. Instead, the intact EpCAM ectodomain was recovered in a soluble form. The researchers provided evidence that EpCAM can be cleaved from exosomes via serum metalloproteinases and indicated that loss of EpCAM on serum exosomes may hamper tumor-derived enrichment of exosomes from blood by immune-affinity isolation.
In the last 5 years, numerous laboratories have begun to intensively advance techniques to isolate pure exosome subpopulations. In this regard, Kim et al. (49) developed zwitterionic polymer-coated immunoaffinity beads to reduce nonspecific protein adsorption from serum for diagnostic applications of exosomes, employing a zwitterionic sulfobetaine monomer with an amine functional group. They coated the beads with poly acrylic acids (PAA) to increase bio-recognition sites, and then, conjugated protein G with carboxylic acid groups on the surfaces for controlling EpCAM antibody orientation. The remaining free carboxylic acid groups were modified with sulfobetaine moieties. These novel beads could successively suppress nonspecific protein adsorption and effectively capture the target protein EpCAM on the exosome surface. Im et al. (50) designed a new surface plasmon resonance chip, named nano-plasmonic exosome (nPLEX) sensor that comprises arrays of periodic nanoholes patterned in a metal film. Each array was functionalized with affinity ligands for different exosomal protein markers. The researchers compared ascites samples from ovarian cancer patients with non-cancerous ascites from cirrhosis patients as controls. Whereas the concentrations of (normal) exosomes, estimated by nPLEX and using CD63 signal changes, could not differentiate between cancer patients and control subjects, the levels of EpCAM- and CD24-positive (tumor-derived) exosomes were significantly higher in ovarian cancer patient samples than in control samples. For electrochemical analysis of the captured exosomes in serum of prostate cancer patients, Zhou et al. (51) developed a microfabricated chip with multiplexed gold sensors. To simultaneously reveal the presence of two specific surface markers on exosomes, electro-oxidation of metal nanoparticles was applied. Silver and copper nanoparticles were used to account for EpCAM and PSMA (prostate-specific membrane antigen), a biomarker enriched on exosomes from prostate cancer cells, respectively. The scientists demonstrated a significant increase in the levels of both EpCAM- and PSMA-positive exosomes in prostate cancer patients compared with healthy men. The high sensitivity of the electrochemical assay was warranted by a detection limit of 50 exosomes per sensor. Mizutani et al. (52) also extracted PSMA-positive exosomes, but incubated plasma of prostate cancer patients with PSMA-conjugated Dynabeads. The laboratory found that patients with aggressive prostate cancer exhibited the highest levels of cancer-related exosomes in blood. Zhao et al. (53) fabricated a microfluidic chip using the PDMS (polydimethylsiloxane) base with a curing agent over a master wafer and, then, bound it with a microscope glass slide. They employed this ExoSearch chip for plasma-based diagnosis of ovarian cancer patients by multiplexed measurement of three exosomal tumor markers (EpCAM, CD24 and CA125). Likewise, Fang et al. (54) developed a microfluidic device composed of a glass substrate and a PDMS membrane that enable on-chip immunocapture exosomes from both cell culture medium and patient plasma. Antibodies against EpCAM and HER2 and an inverted fluorescent microscope were used to visualize the tumor-derived exosomes. The authors demonstrated that the average levels of on-chip captured EpCAM-positive exosomes from breast cancer patients were significantly higher than those of the healthy women, and that the expression levels of exosomal HER2 were almost consistent with those in tumor tissues assessed by immunohistochemical staining. Vaidyanathan et al. (55) developed a multiplexed microfluidic device for specifically capturing multiple exosome targets using a tunable alternating current electrohydrodynamic (ac-EHD) methodology, referred to as nanoshearing. In their system, electrical body forces generated by ac-EHD act within nanometers of an electrode surface, to generate nanoscaled fluid flow that enhances the specificity of exosome capturing and also reduces nonspecific adsorption of weakly bound molecules on the electrode surface. This device exhibited a three-fold enhancement in detection sensitivity in comparison with hydrodynamic flow-based assays. The researchers performed experiments on this device using serum from HER2-positive and -negative breast cancer patients. The samples were driven by individual channels that were functionalized with anti-HER2-specific antibodies. Colorimetric readouts displayed high capture capacity in the HER2-positive patients, whereas negligible capacity levels were observed in HER2-negative patients. To verify the selectivity and specificity of exosome capturing, the patient serum samples were also driven through an anti-CD9 functionalized device. In this case, the capture performance was found to be almost similar for both patient cohorts.
A relatively new method is the nanoparticle tracking analysis (NTA), to measure the size distribution of exosomes. Zhang et al. (56) measured the EpCAM expression on exosomes by NTA. In a fluorescent mode, they conjugated quantum dots with anti-EpCAM to label exosomes for the detection by NTA. They found that exosomes derived from cancer were significantly smaller than those derived from normal cells. Besides, exosomes from different tumor cell lines varied in size, indicating that NTA is an efficient tool for the study on the properties of exosomes in cancer (56). Moreover, Oosthuyzen et al. (57) even applied NTA to urine from healthy volunteers and identified particles with a range of sizes. Using antibodies against the exosome markers CD24 and water channel aquaporin 2 (AQP2) that were conjugated to a fluorophore, they characterized subpopulations of CD24-positive and AQP2-positive particles as exosomes.
All these different technical platforms, here introduced, may extract exosome with high specificity, but some challenges remain to isolate them in sufficient quantity at high purity. Since exosome functions are mainly depending on their functional cargo, e.g., on the content of miRNAs, it is of interest also to examine whether the miRNA profiles in exosomes extracted by the diverse techniques are congruent using the same plasma samples. Thus, a comparison of the different experimental designs is indispensable, to obtain reliable exosomal miRNA profiles specific for the diverse cancer entities, and finally, for a future consensus on a standard operating procedure.
A potential technical paradigm for the fractionation of miRNAs from plasma/serum, tumor-derived exosomes and normal exosomes
The above described technical platforms can be combined to create three fractions for the extraction of cell-free miRNAs from plasma/serum, and exosomal miRNAs from normal and tumor-derived exosomes. In this regard, the protocol for the fractionation contains three steps: extraction of total exosomes from plasma or serum, separation of the exosomes in cancer-derived and normal exosomes, and finally extraction of miRNAs from the exosome subpopulations and the exosome-free plasma/serum supernatant. At first, total exosomes are precipitated in plasma/serum by ultracentrifugation or polymeric methods, e.g., ExoQuick. The pellet contains exosomes from diverse sources, while the supernatant contains cell-free miRNAs that can be extracted by the different techniques, as described above. To verify that the supernatant contains cell-free miRNAs devoid of exosomes, Western blot can be carried out using antibodies specific for AGO2 protein which is complexed with miRNAs (positive result), and for an exosomal marker, e.g., CD63 (negative result) (25). The pellet containing total exosomes is, then, fractionated into cancer-derived and normal exosomes using e.g., the streptavidin/biotin staining method. This method is widely used for sorting specific biomolecules by FACS (fluorescence-activated cell sorting), and particularly suited for the enrichment of tumor-associated exosomes from a solution of total exosomes. The procedure is based on the high binding affinity of streptavidin to biotin mediated by the four binding sites of streptavidin for biotin. In this assay, streptavidin dynabeads or magnetic beads are coupled to a primary biotinylated antibody specific for tumor-derived exosome markers, e.g., EpCAM, HER2, CD24, PSMA etc., and subsequently, incubated with the exosome solution. Once the tumor-derived exosomes are captured by and stabilized on the prepared beads, they can directly be stained by a fluorescent dye, to carry out FACS. This staining is based on FITC (fluorescein isothiocyanate) molecules which are conjugated to a protein known to universally bind to the modifications of exosome surface proteins (e.g., glycosylations and carbohydrate additions). Another method is the indirect staining by a secondary antibody conjugated to FITC (or another fluorescence dye, e.g., Alexa), which binds to the primary antibody. The fluorescently labelled exosomes comprising tumor-associated exosomes are flow-sorted and separated from non-fluorescent exosomes containing normal exosomes. The exosomal miRNAs are, then, extracted from both fractions by the above-described techniques. Accordingly, the procedure allows the comparison of the complete plasma/serum miRNA profile derived from different sources, including normal and tumor cells, with those detected in the subpopulations of normal and tumor-derived exosomes, and exosome-free plasma/serum.
Conclusions
Investigations on exosomes have gained increasing attention in recent years, since the molecular content of exosomes represents signatures of nucleic acids, such as miRNAs, from donor cells, reflecting the pathology of disease (26). In the course of a disease or treatment, their genetic profiles may change, and accordingly, provide insight into the exosome biology for monitoring the disease. Of wide interest are tumor-derived exosomes that are assumed to deliver their cargo from the tumor to near or distant normal cells, and thereby, altering the phenotype and functions of the target cells to promote tumor progression. They represent a prominent subpopulation of the blood vesicular content, and reflect the patient’s tumor status. To date, translational studies have focused on the physiologic and pathophysiologic functions of circulating exosomes along with their genetic signatures as a surrogate for liquid tumor biopsy (26). Not only exosomes derived from the primary tumor or metastases may be eligible for cancer personalized diagnostics, but also exosomes derived from other organs that are affected by tumor burden (58). However, essential for their successful implementation in the clinical setting are research efforts that focus on the development of standardized detection techniques that are capable of isolating, subtyping and quantifying exosomes, as well as revealing the pathogenic role of their cargo. Currently, the lack of standardized techniques, as outlined by the variety of assays in the present review article, limits their entry in the clinic. So, far, there is also no consensus on a gold standard with respect to pre-analytical (e.g., blood sample preparation) and analytical (e.g., extraction, quantification) proceedings.
In terms of the enrichment of tumor-derived exosomes, it is important to comprehensively decipher surface proteins eligible for the use as target molecules that are unfortunately poorly defined. The epithelial cell adhesion molecule EpCAM that is frequently used for the detection of CTCs (59), could represent an appropriate marker for capturing tumor-derived exosomes. However, our preliminary data show that low levels of EpCAM-positive exosomes can also be detected in the plasma of healthy individuals. Therefore, to improve the fractionation in EpCAM-positive and -negative exosomes, the establishment of a baseline cut-off-level should be aimed at. Nonetheless, it should be kept in mind that exosomes may shed EpCAM into the blood circulation, and consequently, remain undetectable (48). Furthermore, the cell surface proteoglycan, glypican-1 (GPC1) was reported to be specifically enriched on pancreas cancer-cell-derived exosomes (60), but recently, Lai et al. (61) showed that GPC1 does not appear to be diagnostic for pancreas. In addition, our unpublished data show that pancreas cancer patients and healthy individuals have similar levels of GPC1-positive exosomes. Taking CD24 into consideration as a target molecule for the enrichment, further investigations on this surface protein are required. These findings show that it might be difficult, to find a cancer-specific exosome surface protein not to be expressed in healthy individuals. It seems that commonly low levels of “tumor-related” antigens exist on healthy exosomes.
A further crucial issue that should be addressed following the standardization of the extraction methods of exosomes and exosomal miRNAs is the data normalization. So far, there is no standardized endogenous reference miRNA, to warrant the reliability of miRNA quantification (37). The choice of a steadily expressed normalizer miRNA is of utmost importance, since it can eliminate differences due to sampling and quality of RNA. Finally, the establishment of a robust and sensitive procedure for the extraction and detection of exosomes and exosomal miRNAs requires the investigation of large populations of plasma samples, with similar clinical parameters, across independent laboratories, to ensure the reproducibility of research data.
Thus, a lot of basic lab work will be needed before tumor-derived exosomes can enter the clinic as therapeutic target molecules or cancer biomarkers. Whether the inhibition of exosomal miRNAs or tumor-derived exosomes that also contain a wide range of other molecules besides oncogenic miRNAs is superior for personalized targeted therapy remains to be examined.
Acknowledgments
I am grateful to the Wilhelm Sander Stiftung, München, Germany, for supporting our study on exosome fractionation.
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Translational Cancer Research for the series “Technologies in Liquid Biopsies - Potential applications in Medicine”. The article has undergone external peer review.
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.09.50). The series “Technologies in Liquid Biopsies - Potential applications in Medicine” was commissioned by the editorial office without any funding or sponsorship. The author has no other conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0.
References
- Mandel P, Métais P. Les acides nucléiques du plasma sanguin chez l`homme. C R Acad Sci Paris 1948;142:241-3.
- Vasioukhin V, Anker P, Maurice P, et al. Point mutations of the N-ras gene in the blood plasma DNA of patients with myelodysplastic syndrome or acute myelogenous leukaemia. Br J Haematol 1994;86:774-9. [Crossref] [PubMed]
- Sorenson GD, Pribish DM, Valone FH, et al. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol Biomarkers Prev 1994;3:67-71. [PubMed]
- Gahan PB, Stroun M. The virtosome-a novel cytosolic informative entity and intercellular messenger. Cell Biochem Funct 2010;28:529-38. [Crossref] [PubMed]
- Stroun M, Lyautey J, Lederrey C, et al. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin Chim Acta 2001;313:139-42. [Crossref] [PubMed]
- Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011;11:426-37. [Crossref] [PubMed]
- Schwarzenbach H, Muller V, Milde-Langosch K, et al. Evaluation of cell-free tumour DNA and RNA in patients with breast cancer and benign breast disease. Mol Biosyst 2011;7:2848-54. [Crossref] [PubMed]
- Okada H, Kohanbash G, Lotze MT. MicroRNAs in immune regulation--opportunities for cancer immunotherapy. Int J Biochem Cell Biol 2010;42:1256-61. [Crossref] [PubMed]
- Lawrie CH, Gal S, Dunlop HM, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol 2008;141:672-5. [Crossref] [PubMed]
- Schwarzenbach H, Nishida N, Calin GA, et al. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol 2014;11:145-56. [Crossref] [PubMed]
- Turchinovich A, Weiz L, Langheinz A, et al. Characterization of extracellular circulating microRNA. Nucleic Acids Res 2011;39:7223-33. [Crossref] [PubMed]
- Trams EG, Lauter CJ, Salem N Jr, et al. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim Biophys Acta 1981;645:63-70. [Crossref] [PubMed]
- Valadi H, Ekstrom K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654-9. [Crossref] [PubMed]
- Hannafon BN, Trigoso YD, Calloway CL, et al. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res 2016;18:90. [Crossref] [PubMed]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005;120:15-20. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215-33. [Crossref] [PubMed]
- Höck J, Meister G. The Argonaute protein family. Genome Biol 2008;9:210. [Crossref] [PubMed]
- Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol 2012;19:586-93. [Crossref] [PubMed]
- Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A 2004;101:2999-3004. [Crossref] [PubMed]
- Pant S, Hilton H, Burczynski ME. The multifaceted exosome: biogenesis, role in normal and aberrant cellular function, and frontiers for pharmacological and biomarker opportunities. Biochem Pharmacol 2012;83:1484-94. [Crossref] [PubMed]
- Zhao L, Liu W, Xiao J, et al. The role of exosomes and "exosomal shuttle microRNA" in tumorigenesis and drug resistance. Cancer Lett 2015;356:339-46. [Crossref] [PubMed]
- Igaz I, Igaz P. Tumor surveillance by circulating microRNAs: a hypothesis. Cell Mol Life Sci 2014;71:4081-7. [Crossref] [PubMed]
- Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev 2013;32:623-42. [Crossref] [PubMed]
- Melo SA, Sugimoto H, O'Connell JT, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 2014;26:707-21. [Crossref] [PubMed]
- Meng X, Muller V, Milde-Langosch K, et al. Diagnostic and prognostic relevance of circulating exosomal miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer. Oncotarget 2016;7:16923-35. [Crossref] [PubMed]
- Schwarzenbach H. The clinical relevance of circulating, exosomal miRNAs as biomarkers for cancer. Expert Rev Mol Diagn 2015;15:1159-69. [Crossref] [PubMed]
- Ostenfeld MS, Jeppesen DK, Laurberg JR, et al. Cellular disposal of miR23b by RAB27-dependent exosome release is linked to acquisition of metastatic properties. Cancer Res 2014;74:5758-71. [Crossref] [PubMed]
- Zhou W, Fong MY, Min Y, et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 2014;25:501-15. [Crossref] [PubMed]
- Chen WX, Zhong SL, Ji MH, et al. MicroRNAs delivered by extracellular vesicles: an emerging resistance mechanism for breast cancer. Tumour Biol 2014;35:2883-92. [Crossref] [PubMed]
- Chen WX, Liu XM, Lv MM, et al. Exosomes from drug-resistant breast cancer cells transmit chemoresistance by a horizontal transfer of microRNAs. PLoS One 2014;9:e95240 [Crossref] [PubMed]
- Tiberio P, Callari M, Angeloni V, et al. Challenges in Using Circulating miRNAs as Cancer Biomarkers. Biomed Res Int 2015;2015:731479 [Crossref] [PubMed]
- Koberle V, Kakoschky B, Ibrahim AA, et al. Vesicle-associated microRNAs are released from blood cells on incubation of blood samples. Transl Res 2016;169:40-6. [Crossref] [PubMed]
- Kirschner MB, Kao SC, Edelman JJ, et al. Haemolysis during sample preparation alters microRNA content of plasma. PLoS One 2011;6:e24145 [Crossref] [PubMed]
- Blondal T, Jensby Nielsen S, Baker A, et al. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods 2013;59:S1-6. [Crossref] [PubMed]
- Chen C, Ridzon DA, Broomer AJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 2005;33:e179 [Crossref] [PubMed]
- Chugh P, Dittmer DP. Potential pitfalls in microRNA profiling. Wiley Interdiscip Rev RNA 2012;3:601-16. [Crossref] [PubMed]
- Schwarzenbach H, Machado da Silva A, Calin G, et al. Data Normalization Strategies for MicroRNA Quantification. Clin Chem 2015;61:1333-42. [Crossref] [PubMed]
- Szatanek R, Baj-Krzyworzeka M, Zimoch J, et al. The Methods of Choice for Extracellular Vesicles (EVs) Characterization. Int J Mol Sci 2017;18. [PubMed]
- Grigor'eva AE, Tamkovich SN, Eremina AV, et al. Characteristics of exosomes andmicroparticles discovered in human tears. Biomed Khim 2016;62:99-106. [Crossref] [PubMed]
- Greening DW, Xu R, Ji H, et al. A protocol for exosome isolation and characterization: evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Methods Mol Biol 2015;1295:179-209. [Crossref] [PubMed]
- Andreu Z, Yanez-Mo M. Tetraspanins in extracellular vesicle formation and function. Front Immunol 2014;5:442. [Crossref] [PubMed]
- Kalra H, Adda CG, Liem M, et al. Comparative proteomics evaluation of plasma exosome isolation techniques and assessment of the stability of exosomes in normal human blood plasma. Proteomics 2013;13:3354-64. [Crossref] [PubMed]
- Taylor DD, Zacharias W, Gercel-Taylor C. Exosome isolation for proteomic analyses and RNA profiling. Methods Mol Biol 2011;728:235-46. [Crossref] [PubMed]
- Lai RC, Yeo RW, Lim SK. Mesenchymal stem cell exosomes. Semin Cell Dev Biol 2015;40:82-8. [Crossref] [PubMed]
- Trzpis M, McLaughlin PM, de Leij LM, et al. Epithelial cell adhesion molecule: more than a carcinoma marker and adhesion molecule. Am J Pathol 2007;171:386-95. [Crossref] [PubMed]
- Esmaeilsabzali H, Beischlag TV, Cox ME, et al. Detection and isolation of circulating tumor cells: principles and methods. Biotechnol Adv 2013;31:1063-84. [Crossref] [PubMed]
- Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol 2008;110:13-21. [Crossref] [PubMed]
- Rupp AK, Rupp C, Keller S, et al. Loss of EpCAM expression in breast cancer derived serum exosomes: role of proteolytic cleavage. Gynecol Oncol 2011;122:437-46. [Crossref] [PubMed]
- Kim G, Yoo CE, Kim M, et al. Noble polymeric surface conjugated with zwitterionic moieties and antibodies for the isolation of exosomes from human serum. Bioconjug Chem 2012;23:2114-20. [Crossref] [PubMed]
- Im H, Shao H, Park YI, et al. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat Biotechnol 2014;32:490-5. [Crossref] [PubMed]
- Zhou YG, Mohamadi RM, Poudineh M, et al. Interrogating Circulating Microsomes and Exosomes Using Metal Nanoparticles. Small 2016;12:727-32. [Crossref] [PubMed]
- Mizutani K, Terazawa R, Kameyama K, et al. Isolation of prostate cancer-related exosomes. Anticancer Res 2014;34:3419-23. [PubMed]
- Zhao Z, Yang Y, Zeng Y, et al. A microfluidic ExoSearch chip for multiplexed exosome detection towards blood-based ovarian cancer diagnosis. Lab Chip 2016;16:489-96. [Crossref] [PubMed]
- Fang S, Tian H, Li X, et al. Clinical application of a microfluidic chip for immunocapture and quantification of circulating exosomes to assist breast cancer diagnosis and molecular classification. PLoS One 2017;12:e0175050 [Crossref] [PubMed]
- Vaidyanathan R, Naghibosadat M, Rauf S, et al. Detecting exosomes specifically: a multiplexed device based on alternating current electrohydrodynamic induced nanoshearing. Anal Chem 2014;86:11125-32. [Crossref] [PubMed]
- Zhang W, Peng P, Kuang Y, et al. Characterization of exosomes derived from ovarian cancer cells and normal ovarian epithelial cells by nanoparticle tracking analysis. Tumour Biol 2016;37:4213-21. [Crossref] [PubMed]
- Oosthuyzen W, Sime NE, Ivy JR, et al. Quantification of human urinary exosomes by nanoparticle tracking analysis. J Physiol 2013;591:5833-42. [Crossref] [PubMed]
- Roccaro AM, Sacco A, Maiso P, et al. BM mesenchymal stromal cell-derived exosomes facilitate multiple myeloma progression. J Clin Invest 2013;123:1542-55. [Crossref] [PubMed]
- Wang L, Balasubramanian P, Chen AP, et al. Promise and limits of the CellSearch platform for evaluating pharmacodynamics in circulating tumor cells. Semin Oncol 2016;43:464-75. [Crossref] [PubMed]
- Melo SA, Luecke LB, Kahlert C, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015;523:177-82. [Crossref] [PubMed]
- Lai X, Wang M, McElyea SD, et al. A microRNA signature in circulating exosomes is superior to exosomal glypican-1 levels for diagnosing pancreatic cancer. Cancer Lett 2017;393:86-93. [Crossref] [PubMed]