
Exosomes in bile as potential pancreatobiliary tumor biomarkers
Introduction
Bile plays a fundamental role in the digestive system by emulsifying lipids and facilitating the proper digestion of fats by lipase action in the small intestine (1). It is mainly composed of water, biliary acids as glycolic and taurocholic acids and bile pigments (bilirubin), as well proteins, electrolytes, mucus and various metabolites (2). It’s well known that bile, secreted by the liver, is transported and stored in the gallbladder and then directed through the biliary tract to the intestine (1,3). Many biliary tract pathologies could have their origin or be the result of alterations in the secretion, composition or transport of bile (4). An example of these alterations is gallstone disease which, when detected; has organ extraction as its standard procedure (5). However, the chronic presence of these stones in the epithelial tissue induces chronic inflammation and the potential development of several complications like cancer (6). Therefore, detection of early molecular alterations as biomarkers of biliary tract pathology is clinically important (7). This is useful considering that smaller organs such as the gallbladder and the biliary ducts, are difficult to clinically explore (8).
The term ‘biomarker’ refers to medical signs (including the detection of molecules) that are accurately measurable in a reproducible manner and that account for a pathological state in the patient or a group of them (9). Therefore, biomarkers represent a medical condition that can be detected externally and has a rapid and selective accumulation which accounts for morphological or biochemical alterations (10). However, one of the main aspects that makes a biomarker a good candidate for its use is the technique employed for its detection (9). Considering the above, blood biomarkers that identify certain pathophysiological conditions have been widely used; due to their easy detection and non-invasive acquisition (11). However these biomarkers aren’t exempt of problems (10). In this regard, there are several research works designed to identify biomarkers that account for oncological pathologies in the biliary tract in serum and bile fluid.
Pancreatobiliary pathologies detection
Biliary tract malignancies originate from epithelial cells of the intrahepatic or extrahepatic bile ducts. They comprise intrahepatic cholangiocarcinoma (CCA), perihilar CCA, gallbladder cancer (GBC) and distal CCA. These cancers, present a diagnostic challenge and a conundrum (12), with a very poor outcomes. Surgical resection is the only chance for cure in early stages of the disease (13). If resected, 5 year survival reaches 30–40% for early stage intrahepatic CCA and 50% for extrahepatic CCA, but less than 20% in advanced GBC (14). Unfortunately, in unresectable disease, median overall survival rates are only 6–12 months (15,16). The majority of these tumors are discovered at an advanced stage due to a delay in diagnosis and, currently, the clinical detection and diagnosis relies on computed tomography (CT) or B type ultrasonography examinations, which have poor sensitivity, especially for the detection of small lesions (17–19). Histology combined with cytology by endoscopic ultrasound-guided fine needle aspiration for the diagnosis of solid pancreatic mass including distal biliary tract tumors has become the standard. However is not available in all institutions and is highly invasive (20). In addition, brush cytology via endoscopic retrograde cholangiopancreatography (ERCP) has a sensitivity of 50% for early diagnosis of CCA, which is attributed to the desmoplastic nature of this disease (21). While most bile duct carcinomas develop sporadically, there are some known risk factors, including parasitic infestation, choledochal cysts, hepatitis C virus, intrahepatic lithiasis, abnormal pancreatobiliary junction, and primary sclerosing cholangitis (PSC) (22). PSC carries the highest risk of CCA development (23) and PSC is a frequent early event during cholangiocarcinogenesis. Furthermore, half of patients with PSC that develop cancer will do so within a year of diagnosis (24,25). Despite advances in endoscopy and multiple imaging techniques, all of them lack diagnostic accuracy and/or negative predictive value and large efforts have aimed to identify reliable biomarkers of biliary malignancies that could replace the current clinical gold standards (26,27).
Until now, serum biomarkers lack the sufficient sensitivity and specificity to reliably screen high risk individuals or confirm the presence of malignancy in biliary structures (22,28). The most widely used clinical biomarker for biliary tract malignancies is serum carbohydrate antigen 19-9 (CA19-9) (29). However, CA19-9 may also be elevated in pancreatitis, cholangitis, primary biliary cirrhosis and with heavy tobacco use (30). The range of sensitivity and specificity for CA19-9 is between 53–92% and 50–98%, depending on the cutoff value used and population studied (31). Alternative biomarkers have been identified and investigated (32), but there is no validated biomarker with clinically useful diagnostic capabilities for biliary malignancies to date. Therefore, improved fluid-based biomarkers are urgently required to enable early tumor diagnosis and many efforts have been made to study the bile fluid as a source of diagnostic biomarkers from the biliary tree in health and disease. Bile fluid is in direct contact with the bile duct epithelia, making it an attractive option to investigate molecules with diagnostic and prognostic potential. However, the acquisition of bile fluid through invasive techniques as ERCPs is not an easy way to do so and needs a medical specialist (33), therefore the use for screening purposes or surveillance of population at risk is limited.
Biomarkers and pancreatobiliary tract tumors
CA 19-9 and carcinoembryonic antigen (CEA) are widely used in the clinical practices and the utility of this glycoprotein in bile has been studied too. However diagnostic performance has not been consistent. For example, biliary CEA showed a 84% of sensitivity and a specificity of 64%, but there was a considerable overlap between the malignant and benign lesions (34,35). Furthermore, in the multivariate analysis, biliary CEA levels were not predictive of malignancy (34). The low to moderate specificities for these markers suggest that they are increased in benign and inflammatory conditions (32). Multiple studies have shown that biliary CA 19-9 and CEA did not add to the diagnostic accuracy to a great extent when compared to the serum levels, as they had high false positive results (32,36).
Other bile biomarkers have been investigated as a potential, more sensitive, alternative due to direct contact with tumor. For example, IGF-1 and VEGF were studied in different works as bile diagnostic markers (31,32). IGF-1 was found to be diagnostic for CCA, when benign conditions or pancreatic cancer were taken as a control. Previous studies have demonstrated that biliary IGF-1 was 15–20 folds higher in patients with extrahepatic CCA compared to patients with pancreatic carcinoma or benign disease (37). However, the levels of biliary IGF-1 didn’t correlate with the degree of cholestasis. Increased biliary VEGF levels have been reported in pancreatic cancer compared to CCA, with a sensitivity of 93.3% and a specificity of 88.9%. Also, increased VEGF levels could be differentiating pancreatic cancer from benign lesions (32,38). However, biliary VEGF levels did not differ significantly between the benign and CCA group (37). Several research groups have employed different proteomic analysis to identify novel tumor-specific markers in bile, which carries proteins from the local environment (liver, biliary tract and pancreas) (39,40). Some of the analytical techniques used in proteomics are the liquid chromatography-mass spectrometry and nuclear magnetic resonance spectroscopy, besides Western blot and ELISA. Recently, was reported that the SSP411 protein in bile as a novel and specific biomarker for CCA compared to benign group using the 2D/MS/MS strategy (41). Neutrophil Gelatinase-Associated Lipocalin (NGAL), was suggested as a biomarker for distinguishing benign from malignant pancreatobiliary obstruction, and the combination of biliary NGAL and serum CA19-9 improved diagnostic accuracy for malignancy (42). Carcinoembryonic Cell Adhesion Molecule 6 (CEAM6) in bile was described as a useful biomarker to discriminate between malignant and nonmalignant causes of biliary stricture (43). Interestingly, a panel comprised by CEAM6 and serum CA19-9 further improved diagnostic accuracy for malignant stenoses (43). Recently, the Alpha-1-antitrypsin was reported as an overexpressed protein that’s increased in CCA bile samples and could differentiate between normal and early-stage CCA (44). Interestingly, in this work, CEAM6 was not increased between normal and malignant samples, suggesting that differences in sample number and analytical techniques could affect the performance of the same biomarker. Furthermore, the main limitation of proteomic analysis of bile is its complex constitution with various molecular components and contaminants. Therefore, a variety of sample preparations, including delipidation, desalination and nucleic acid removal, must be adopted to remove interfering substances. Also, proteins account for approximately 7% of the total dry weight; and differential fractionation could be used to reduce the mix complexity, concentrating the protein component as a preparation for mass spectrometry (45,46).
In recent years, metabolomics analysis has emerged as an effective tool for the screening of biomarkers and disease diagnosis. For example, was demonstrated a clear separation between the disease groups (biliary tract cancer and benign biliary tract diseases) in bile samples through a liquid chromatography/mass spectrometry (LC/MS)-based approach (47). Patients with biliary tract cancer showed significantly lower levels of lysophosphatidylcholine, phenylalanine, 2-octenoylcarnitine, tryptophan and significantly higher levels of taurine and glycine conjugated bile acids in the bile compared with those in the bile from patients with benign biliary tract disease (47). These metabolites may have potential as novel biomarkers for the early detection of biliary tract cancer and further studies need to be done to validate their individual clinical applicability. However, the technological limitations in the implementation of these methods and the necessary expertise training of the personnel make these analyses impractical to use in the clinical diagnostic routine.
Detection of mutated genes in bile might be another approach for early detection of pancreatobiliary tract carcinoma and different groups have reported the detection of different mutation derived from transformed cells in bile compared to clinical samples obtained from individuals with benign pancreatobiliary disorders. For example, the detection of codon 12 mutation of KRAS in bile fluid of PSC patients (48). Interestingly, most of the PSC patients with KRAS mutations remained tumor free after a follow up, which agrees with the fact that these mutations are not specific for malignancy but may also occur in normal bile duct mucosa or in dysplasia (48). In this scenario, the KRAS codon 12 mutation in bile fluid appears to be a prognostic factor for PSC patients. Others groups analyzed different mutant KRAS2 genes by LigAmp assay in bile (49). Data showed the presence of mutant KRAS2 in bile in the majority of the cancer patients (87.5%) and in a minor subset of benign pancreatobiliary diseases (49). Therefore, analysis of KRAS2 mutations in bile could be a sensitive assay for an early detection of biliary tract carcinoma. Other group used a different approach, the analysis of a DNA methylation marker panel for the detection of extrahepatic cholangiocarcinoma (EHC) in bile (50). In this work, a five-gene panel (CCND2, CDH13, GRIN2B, RUNX3, and TWIST1) detected EHC at a sensitivity of 83%, which was far higher than that of bile cytology (46%). However, the authors recommended validating the usefulness of this test by performing large-scale or multicenter studies, because the control sample size was small (50). The detection of RNA as potential biomarker also was explored as a potential biomarker of CCA. Used U2 small nuclear RNA fragments (RNU2-1f) in bile enabled the differentiation of patients with CCA from control samples (patients with PSC and choledocholithiasis) (51). However, there was some overlap in the RNU2-1f abundance between the CCA and PSC samples, showing a lower sensitivity and specificity (67% and 91%, respectively) (51).
Extensive evidence indicates that most cancers exhibit alterations related to the expression of microRNAs (miRNAs) and tumor cells could express high levels of pro-carcinogenic miRNAs or lower levels of tumor suppressor miRNAs (52). High levels of miR-21, miR-187 and miR-202 in blood can be used to diagnose GBC (53). Also, lower levels of miR-143, Let-7a and miR-335 are found in serum and tissue of GBC patients with respect to healthy controls. Interestingly, miR-143, miR-202 and miR-187 showed direct association in blood, tissues and in lymphatic metastasis, and may be useful for diagnosis, treatment and monitoring of therapies. miR-150 could be used as a biomarker for CCA, since it is elevated in serum of patients with this disease (54). However, the main disadvantage of the use of miRNAs as biomarkers in systemic extracellular fluids is the likely effects of extracellular enzymes such as serum nucleases, which degrade miRNA molecules. Lately, some groups have described the miRNA detection in bile as a biomarker of pancreatobiliary disease. An study demonstrated that 10 of the 667 bile miRNAs analyzed (miR-9, miR-145, miR-105, miR-147b, let-7f-2,let-7i, miR-302c, miR-199a-3p, miR-222 and miR-942) were expressed significantly higher in the malignant group, composed of CCA and GBC patients than in the control group (patients with choledocholithiasis) (55). Also, patients with PSC and CCA, have distinct miRNA profiles in bile (56). miR-412, miR-640, miR-1537 and miR-3189 expression was different between patients with PSC and PSC/CCA in bile samples of which only miR-412 was upregulated.
However, one of the main difficulties detecting and analyzing free nucleic acids with diagnostic value in bile is the presence of multiple PCR inhibitors. Iron-containing proteins and their breakdown products, such as bilirubin, and bile salts have been identified as major inhibitors in PCR (57). Further studies are needed to determine ways to minimize the inhibitory effects of bile components in PCR and thus improve the detection sensitivity.
Moreover, the evidence suggests that the diagnostic use of a biomarker could be complemented by other biomarkers that account for the same pathology or clinical stage of disease. However, the large number of molecules in the serum and the limited specificity that these biomarkers offer can represent a disadvantage (58). The latter, because the blood is a tissue that delivers and collects substances at the systemic level, while the composition of other body fluids, such as bile, may be more representative of a localized body area, which would result in an advantage when evaluating biomarkers of the biliary tract under pathophysiological conditions. These represents a challenge in the field of prevention, diagnosis, prognosis and monitoring of oncological or non-oncological pathologies of the biliary tract (41,59). In this context, the use of exosomes could be key to the successful management of pathologies difficult to explore, such as those related to the biliary tract (60,61).
Extracellular microvesicles
Extracellular vesicles (EVs) have become an interesting focus of research due to their functions both in physiological and pathological conditions (62). Cells can generate different types of these EVs, which are released directly from the plasma membrane (63). These vesicles are classified by its size and biogenesis as apoptotic bodies, microvesicles and exosomes, among others. Accordingly, exosomes are homogeneous vesicles (50–150 nm), composed of a lipid bilayer that originates from multivesicular bodies (MVBs) and represent a discrete subpopulation of EVs (63,64). Exosomes have received tremendous attention in recent years since its content mirrors the parental cells they are originated from (65). Although the exact biological functions of exosomes remain to be fully uncovered, current evidence indicates that exosomes play a vital role in many cellular processes like cell-cell communication, antigen presentation, coagulation, waste management, as well as protein, DNA and RNAs transfer (66,67). Thus, the EVs are considered potential messengers of information between distant cells (68). Therefore, the detection and analysis of these EVs as biomarkers in several diseases or their use as vehicles for different types of therapies have been of great interest in recent years. Interestingly, an important characteristic of the exosomes, and useful to use as molecular biomarker, is the ability to transport and protect nucleic acids or protein content from extracellular enzymes and contaminants (69). For that reason, circulating exosomes provide a promising approach as novel and dynamic biomarkers in human diseases due to their stability, accessibility and molecular representation of the cells that secrete them. The latter takes relevance if we consider that tumor-derived exosomes are found in all body fluids, such as blood, urine, saliva, pleural effusions and malignant ascite, reflecting the molecular changes during the early events of many pathologies including cancer (61,70). In recent years, the isolation and quantification of exosomes have become a major initiative in both basic research and clinical application (71), inspiring different research groups and biotechnological companies to develop techniques and protocols to reliably and efficiently isolate exosomes from complex biological matrices (72).
Unfortunately, circulating fluids reflect all pathophysiological changes that occur within the body and quantitative and qualitative changes of EVs could arise due to other conditions (73). Therefore, to preserve the diagnostic relevance, non-body circulating fluids as bile, could represent a valuable alternative source for EVs like exosomes, because they remain virtually unaffected by the different processes that occur in distant organs.
Exosomes and pancreatobiliary tract tumors
Several studies have shown that tumor cells produce and release exosomes in greater quantity than normal cells, transmitting messages to normal cells or other transformed cells at nearby or distant sites, promoting tumor growth and progression (74). Exosomes contribute to cancer proliferation by supplying different anti-apoptotic proteins, miRNAs, oncogenic proteins, cytokines, adhesion molecules among others (75). Moreover, exosomes derived from different types of cancer as glioblastoma, breast cancer and multiple myeloma are demonstrated to upregulate angiogenesis by the transmission of miRNAs or angiogenic proteins (76).
The presence and biological importance of EVs as exosomes in the pancreatobiliary tract and its function depending of cellular origin and the physiological and pathophysiological context in which they are secreted have only been recently studied. Exosomes derived from pancreatic ductal adenocarcinoma cells, released into the blood circulation have been shown to promote a favorable fibrotic microenvironment in the liver, a pre-metastatic niche, supporting metastasis formation in a mouse model (77).
Exosomes produced by aggressive CCA cell lines transferred oncogenic proteins and induced migration and invasion of normal human cholangiocyte (H69) cells, suggesting a direct role of these microvesicles in cell to cell communication and the facilitation of a tumor permissive microenvironment (78). Also, exosomes from malignant cholangiocytes can increase fibroblast-like activity by mesenchymal stem cells (MSCs) (79). Furthermore, exposure of MSCs to tumor cell–derived EVs results in a selective alteration of mRNA expression and release of cytokines/chemokines such as IL-6 that can, in turn, alter tumor cell growth. Also, exosomes derived from CCA cells has immunomodulatory properties (80). These exosomes inhibit the antitumor activity of CIK cells by down-regulating the population of CD3+, CD8+, NK (CD56+), and CD3+CD56+ cells and the secretion of TNF-α and perforin, highlighting the importance of exosomes in the interrelationship of tumor and microenvironment, facilitating the cancer progression.
Exosomes as pancreatobiliary tract tumors biomarkers
Several research groups have proposed exosomes as a potential screening biomarkers or therapeutic targets in biliary tract diseases (Table 1). On this subject, different studies have related the presence and secretion of EVs with an altered expression of different molecules with diagnostic potential. Recently, were described that concentration of serum EVs in HCC samples was higher than the other study groups (Control, CCA and PSC) (91). Also, through a proteomic analysis of exosomes, was identifying specific proteins with potential diagnostic and prognostic value for CCA, PSC and HCC (91). Moreover, exosomes derived from pancreatic ductal adenocarcinoma patients could be a source of DNA allowing the identification of mutations in the KRAS gene and pointing out the utility of the analyses of these EVs as noninvasive screening methods (87).
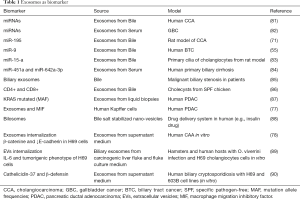
Full table
The first evidences of the physiological role of EVs in biliary tract were given by the identification of EVs in bile (83). Bile EVs were partially secreted by cholangiocytes and directly bound to their primary cilium inhibiting cell proliferation in an ERK-dependent manner which is associated with lower levels of miRNA 15-A (83). These data evidenced that EVs may contribute to the maintenance of the homeostasis of the biliary epithelia in normal conditions. EVs present in chicken bile induce an increase in the proliferation of CD4+ and CD8+ T lymphocytes, they promote the activation of liver macrophages and the inhibition of the replication of avian leukosis virus in a cell line of chicken fibroblasts (DF-1 cells) (86). Furthermore, in other pathologies associated with the biliary tract, it’s known that these vesicles may play an important role in promoting an immune reaction in biliary infections, as in the case of infection with Cryptosporidium parvum (90). This parasite induces the release of vesicles loaded with antimicrobial peptides, such as cateliadine-37 and beta-defensin 2, when interacting with the TLR4/IKK2 receptor of the bile epithelium (90).
EVs are present in human bile and contain a large number of miRNA making these molecules very stable (81). A group of miRNAs were described to be upregulated in EVs isolated from bile of CCA patients compared to several biliary benign disorders (i.e., choledocholithiasis, PSC, chronic pancreatitis and Sphincter of Oddi dysfunction) showing a diagnostic value (81). Interestingly, combining five miRNA markers (miR-191, miR-486-3p, miR-1274b, miR-16 and miR-484) could differentiate between CCA from PSC and other bile duct obstruction, with a sensitivity of 67% and a specificity of 96%. A recent work, pointing out the quantity of biliary exosomes as markers of malignant biliary stenosis, which may be useful in replacing CA19-9 antigen detection in serum (85). This work, indicated that the median concentration of EVs was significantly higher in bile samples from patients with malignant than controls or nonmalignant common bile duct stenoses, and the concentration of EVs in bile samples could discriminate between patients with a 100% of accuracy. Better than the concentration of EVs in serum 63.3% diagnostic accuracy.
Finally, EVs as exosomes have been recognized as a valuable resource for the detection and monitoring of cancer, being shown as a tool of non-invasive detection in this and other type of pathologies. The latter is due to its content and molecular nature, which reflects the pathophysiological state of the cells that secrete them (65). Unfortunately, few studies are focused in the diagnostic potential of EVs as exosomes in bile, due to the difficulties associated to the acquisition of sample, the lack of standardized protocols for exosomes isolation, and the gap between the basic and clinical research to validate different molecules with potential diagnostic or prognostic value better than the available serum gold standard biomarkers.
Concluding remarks
The diagnostic of biliary structures alterations represent a medical challenge, also the initial workup, as abdominal imaging and ERCP based sampling, is no diagnostic (32,92). Regrettably, the majority of biliary tract tumors are discovered at an advanced stage due to a delay in diagnosis and the overall survival of these patients is dismal. Current diagnosis methods are based on a combination of radiological imaging, serum markers and histological verification; however, each of these approaches has its own drawback. Therefore, is an urgent need to create new, better and accurate diagnostic methods to detect early-stage tumors in high-risk groups (Table 2). Although both CA19-9 and CEA are elevated in the serum of patients with biliary tract malignancies, these markers are also increased in other diseases such as alcoholic liver disease, viral hepatitis, PSC, cholestasis, liver injury, and other tumors. Thus, the diagnostic value of these serum biomarkers is limited.
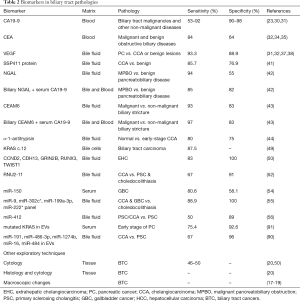
Full table
In recent years, several research and technological advances have been made in the field of liquid biopsy, a new approach to detect and characterize genetic alterations derived from tumors in systemic fluids such as blood or other restricted fluids such as cerebrospinal fluid, avoiding the need for surgery for acquisition of tissue. In addition, the development and enhancement of several technology platforms such as qPCR gene panels, target sequencing, digital PCR, capture of circulating tumor cells (CTC) and others allow the detection of known genetic alterations or the identification of new relevant molecular changes for the disease, improving the diagnostic, prognostic and predictive value of these new biomarkers with several advantages in sensitivity and specificity. Interestingly, the proximity of bile fluid to the biliary epithelia makes it an attractive option for researching new molecular biomarkers, which might be representative of the abnormal changes taking place in the biliary system and minimal or non-influenced by whole body changes or other alterations that could affect the serum markers. Bile fluids has recently been proven to contain EVs as exosomes that play a significant role in the pathophysiology of biliary tract diseases becoming in promising non-invasive tools for diagnosis, prognosis and to predict treatment response. The main advantage for the use of biliary EVs as a source of biomolecular diagnostic markers is their intrinsic nature that aims to protect the molecular content from the adverse environment as the bile fluid (Figure 1). The isolation and concentration of EVs from bile could allow identification of proteins and different nucleic acids from tumor cells, without the need of pre-treatment for clearing and removing the contaminants that affect the detection of free biomolecules in bile. Biliary EV and its content showed high sensitivity and specificity for the diagnosis of different biliary tract diseases and its extensive use could be a promising future option as the first diagnostic step in early detection in individuals with known risk factors (i.e., liver flukes, hepatitis B and C or PSC) (13,93).
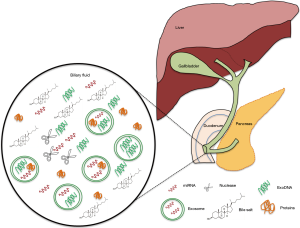
However, it is necessary to point out the need to standardize the isolation, purification and characterization methods for this material and validate its uses as biomarker in large and defined clinical cohorts.
Future perspective
EVs in bile are very promising as biomarkers to distinguish between benign and malignant biliary conditions. However, it will be necessary to identify additional promising biomarkers and validate their usefulness by performing large-scale or multicenter studies. Moreover, special attention should be directed to bridge the gap between pre-clinical and clinical studies. Thus, future international collaborative investigations are urgently needed to validate the most promising biomarkers and to start their implementation in the general clinical practice. On the other hand, the acquisition of bile fluid through invasive technique as ERCPs it’s not an easy way to achieve this and needs medical specialists, therefore the use for screening purposes or surveillance of population at risk is limited.
Acknowledgments
Funding: This work was supported by Fondo Nacional de Desarrollo Científico y Tecnológico (FONDECYT) 1170893; Comisión Nacional de Investigación Científica y Tecnológica (CONICYT) and Instituto Milenio en Inmunología e Inmunoterapia (IMII) P09/016-F.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Edward R. Sauter) for the series “Body Fluid Exosomes and Cancer” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.10.37). The series “Body Fluid Exosomes and Cancer” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Marin JJ, Macias RI, Briz O, et al. Bile Acids in Physiology, Pathology and Pharmacology. Curr Drug Metab 2015;17:4-29. [Crossref] [PubMed]
- Pasternak A, Szura M, Gil K, et al. Metabolism of bile with respect to etiology of gallstone disease - systematic review. Folia Med Cracov 2014;54:5-16. [PubMed]
- Keplinger KM, Bloomston M. Anatomy and embryology of the biliary tract. Surg. Clin. North Am. 2014;94:203-17. [Crossref] [PubMed]
- Schirmer BD, Winters KL, Edlich RF. Cholelithiasis and Cholecystitis. J Long Term Eff Med Implants 2005;15:329-38. [Crossref] [PubMed]
- Williams EJ, Green J, Beckingham I, et al. Guidelines on the management of common bile duct stones (CBDS). Gut 2008;57:1004-21. [Crossref] [PubMed]
- Espinoza JA, Bizama C, García P, et al. The inflammatory inception of gallbladder cancer. Biochim Biophys Acta 2016;1865:245-54. [PubMed]
- Bartley AN, Hamilton SR. Select biomarkers for tumors of the gastrointestinal tract: Present and future. Arch Pathol Lab Med 2015;139:457-68. [Crossref] [PubMed]
- Roa I, Araya JC, Villaseca M, et al. Gallbladder cancer in a high risk area: Morphological features and spread patterns. Hepatogastroenterology 1999;46:1540-6. [PubMed]
- Strimbu K, Tavel JA. What are Biomarkers? Curr Opin HIV AIDS 2010;5:463-6. [Crossref] [PubMed]
- Mayeux R. Biomarkers: potential uses and limitations. NeuroRx 2004;1:182-8. [Crossref] [PubMed]
- Jokerst JV, Gambhir SS. Molecular imaging with theranostic nanoparticles. Acc Chem Res 2011;44:1050-60. [Crossref] [PubMed]
- Hennedige TP, Neo WT, Venkatesh SK. Imaging of malignancies of the biliary tract- an update. Cancer Imaging 2014;14:14. [PubMed]
- Patel T. Cholangiocarcinoma--controversies and challenges. Nat Rev Gastroenterol Hepatol 2011;8:189-200. [Crossref] [PubMed]
- Roa I, Ibacache G, Muñoz S, et al. Gallbladder cancer in Chile: Pathologic characteristics of survival and prognostic factors: Analysis of 1,366 cases. Am J Clin Pathol 2014;141:675-82. [Crossref] [PubMed]
- Blechacz B, Komuta M, Roskams T, et al. Clinical diagnosis and staging of cholangiocarcinoma. Nat Rev Gastroenterol Hepatol 2011;8:512-22. [Crossref] [PubMed]
- Friman S. Cholangiocarcinoma — Current Treatment Options. Scand J Surg 2011;100:30-4. [Crossref] [PubMed]
- Lee ES, Lee JM. Imaging diagnosis of pancreatic cancer: A state-of-the-art review. World J Gastroenterol 2014;20:7864-77. [Crossref] [PubMed]
- Attwa MH, El-Etreby SA. Guide for diagnosis and treatment of hepatocellular carcinoma. World J Hepatol 2015;7:1632-51. [Crossref] [PubMed]
- Razumilava N, Gores GJ. Cholangiocarcinoma. Lancet 2014;383:2168-79. [Crossref] [PubMed]
- Kim TH, Choi KH, Song HS, et al. Histology combined with cytology by endoscopic ultrasound-guided fine needle aspiration for the diagnosis of solid pancreatic mass and intra-abdominal lymphadenopathy. Gut Liver 2013;7:605-10. [Crossref] [PubMed]
- Navaneethan U, Njei B, Lourdusamy V, et al. Comparative effectiveness of biliary brush cytology and intraductal biopsy for detection of malignant biliary strictures: A systematic review and meta-analysis. Gastrointest Endosc 2015;81:168-76. [Crossref] [PubMed]
- Rose JB, Correa-Gallego C, Li Y, et al. The role of biliary carcinoembryonic antigen-related cellular adhesion molecule 6 (CEACAM6) a sa biomarker in cholangiocarcinoma. PLoS One 2016;11:e0150195 [Crossref] [PubMed]
- Lazaridis KN, Gores GJ. Primary sclerosing cholangitis and cholangiocarcinoma. Semin Liver Dis 2006;26:42-51. [Crossref] [PubMed]
- Timmer MR, Lau CT, Meijer SL, et al. Genetic Abnormalities in Biliary Brush Samples for Distinguishing Cholangiocarcinoma from Benign Strictures in Primary Sclerosing Cholangitis. Gastroenterol Res Pract 2016;2016:4381513 [PubMed]
- Singh S, Talwalkar JA. Primary sclerosing cholangitis: diagnosis, prognosis, and management. Clin Gastroenterol Hepatol 2013;11:898-907. [Crossref] [PubMed]
- Aljiffry M, Renfrew PD, Walsh MJ, et al. Analytical review of diagnosis and treatment strategies for dominant bile duct strictures in patients with primary sclerosing cholangitis. HPB (Oxford) 2011;13:79-90. [Crossref] [PubMed]
- Karlsen TH, Folseraas T, Thorburn D, et al. Primary sclerosing cholangitis - a comprehensive review. J Hepatol 2017;17:32196-7. [PubMed]
- Xu MM, Sethi A. Diagnosing Biliary Malignancy. Gastrointest Endosc Clin N Am 2015;25:677-90. [Crossref] [PubMed]
- Malaguarnera G. Markers of bile duct tumors. World J Gastrointest Oncol 2011;3:49. [Crossref] [PubMed]
- Gatto M, Bragazzi MC, Semeraro R, et al. Cholangiocarcinoma: Update and future perspectives. Dig Liver Dis 2010;42:253-60. [Crossref] [PubMed]
- Alvaro D. Serum and bile biomarkers for cholangiocarcinoma. Curr Opin Gastroenterol 2009;25:279-84. [Crossref] [PubMed]
- Lourdusamy V, Tharian B, Navaneethan U. Biomarkers in bile-complementing advanced endoscopic imaging in the diagnosis of indeterminate biliary strictures. World J Gastrointest Endosc 2015;7:308-17. [PubMed]
- Uppal DS, Wang AY. Advances in endoscopic retrograde cholangiopancreatography for the treatment of cholangiocarcinoma. World J Gastrointest Endosc 2015;7:675-87. [PubMed]
- Buffet C, Fourre C, Altman C, et al. Bile levels of carcino-embryonic antigen in patients with hepatopancreatobiliary disease. Eur J Gastroenterol Hepatol 1996;8:131-4. [Crossref] [PubMed]
- Akdoğan M, Parlak E, Kayhan B, et al. Are serum and biliary carcinoembryonic antigen and carbohydrate antigen19-9 determinations reliable for differentiation between benign and malignant biliary disease? Turk J Gastroenterol 2003;14:181-4. [PubMed]
- Lindberg B, Arnelo U, Bergquist A, et al. Diagnosis of biliary strictures in conjunction with endoscopic retrograde cholangiopancreaticography, with special reference to patients with primary sclerosing cholangitis. Endoscopy 2002;34:909-16. [Crossref] [PubMed]
- Alvaro D, Macarri G, Mancino MG, et al. Serum and biliary insulin-like growth factor I and vascular endothelial growth factor in determining the cause of obstructive cholestasis. Ann Intern Med 2007;147:451-9. [Crossref] [PubMed]
- Navaneethan U, Gutierrez NG, Jegadeesan R, et al. Vascular endothelial growth factor levels in bile distinguishes pancreatic cancer from other etiologies of biliary stricture: A pilot study. Dig Dis Sci 2013;58:2986-92. [Crossref] [PubMed]
- Barbhuiya MA, Sahasrabuddhe NA, Pinto SM, et al. Comprehensive proteomic analysis of human bile. Proteomics 2011;11:4443-53. [Crossref] [PubMed]
- Lukic N, Visentin R, Delhaye M, et al. An integrated approach for comparative proteomic analysis of human bile reveals overexpressed cancer-associated proteins in malignant biliary stenosis. Biochim Biophys Acta 2014;1844:1026-33. [Crossref] [PubMed]
- Shen J, Wang W, Wu J, et al. Comparative Proteomic Profiling of Human Bile Reveals SSP411 as a Novel Biomarker of Cholangiocarcinoma. PLoS One 2012;7:e47476 [Crossref] [PubMed]
- Zabron AA, Horneffer-van der Sluis VM, Wadsworth CA, et al. Elevated Levels of Neutrophil Gelatinase-Associated Lipocalin in Bile From Patients With Malignant Pancreatobiliary Disease. Am J Gastroenterol 2011;106:1711-7. [Crossref] [PubMed]
- Farina A, Dumonceau JM, Antinori P, et al. Bile carcinoembryonic cell adhesion molecule 6 (CEAM6) as a biomarker of malignant biliary stenoses. Biochim Biophys Acta 2014;1844:1018-25. [Crossref] [PubMed]
- Laohaviroj M, Potriquet J, Jia X, et al. A comparative proteomic analysis of bile for biomarkers of cholangiocarcinoma. Tumour Biol 2017;39:1010428317705764 [Crossref] [PubMed]
- Farina A, Dumonceau JM, Delhaye M, et al. A step further in the analysis of human bile proteome. J Proteome Res 2011;10:2047-63. [Crossref] [PubMed]
- Bonney GK, Craven RA, Prasad R, et al. Circulating markers of biliary malignancy: opportunities in proteomics? Lancet Oncol 2008;9:149-58. [Crossref] [PubMed]
- Xu X, Cheng S, Ding C, et al. Identification of bile biomarkers of biliary tract cancer through a liquid chromatography/mass spectrometry-based metabolomic method. Mol Med Rep 2015;11:2191-8. [Crossref] [PubMed]
- Kubicka S, Kühnel F, Flemming P, et al. K-ras mutations in the bile of patients with primary sclerosing cholangitis. Gut 2001;48:403-8. [Crossref] [PubMed]
- Shi C, Chandrasekaran A, Thuluvath PJ, et al. Ultrasensitive detection of KRAS2 mutations in bile and serum from patients with biliary tract carcinoma using LigAmp technology. J Mol Diagn 2009;11:583-9. [Crossref] [PubMed]
- Shin SH, Lee K, Kim BH, et al. Bile-based detection of extrahepatic cholangiocarcinoma with quantitative DNA methylation markers and its high sensitivity. J Mol Diagn 2012;14:256-63. [Crossref] [PubMed]
- Baraniskin A, Nöpel-Dünnebacke S, Schumacher B, et al. Analysis of U2 small nuclear RNA fragments in the bile differentiates cholangiocarcinoma from primary sclerosing cholangitis and other benign biliary disorders. Dig Dis Sci 2014;59:1436-41. [Crossref] [PubMed]
- Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol 2012;6:590-610. [Crossref] [PubMed]
- Li G, Pu Y. MicroRNA signatures in total peripheral blood of gallbladder cancer patients. Tumour Biol 2015;36:6985-90. [Crossref] [PubMed]
- Wang S, Yin J, Li T, et al. Upregulated circulating miR-150 is associated with the risk of intrahepatic cholangiocarcinoma. Oncol Rep 2015;33:819-25. [Crossref] [PubMed]
- Shigehara K, Yokomuro S, Ishibashi O, et al. Real-time PCR-based analysis of the human bile micrornaome identifies miR-9 as a potential diagnostic biomarker for biliary tract cancer. PLoS One 2011;6:e23584 [Crossref] [PubMed]
- Voigtländer T, Gupta SK, Thum S, et al. MicroRNAs in serum and bile of patients with primary sclerosing cholangitis and/or cholangiocarcinoma. PLoS One 2015;10:e0139305 [Crossref] [PubMed]
- Schrader C, Schielke A, Ellerbroek L, et al. PCR inhibitors - occurrence, properties and removal. J Appl Microbiol 2012;113:1014-26. [Crossref] [PubMed]
- Sahab ZJ, Semaan SM, Sang QX. Methodology and applications of disease biomarker identification in human serum. Biomark Insights 2007;2:21-43. [PubMed]
- Koopmann J, Thuluvath PJ, Zahurak ML, et al. Mac-2-binding protein is a diagnostic marker for biliary tract carcinoma. Cancer 2004;101:1609-15. [Crossref] [PubMed]
- Properzi F, Logozzi M, Fais S. Exosomes: the future of biomarkers in medicine. Biomark Med 2013;7:769-78. [Crossref] [PubMed]
- Lin J, Li J, Huang B, et al. Exosomes: Novel Biomarkers for Clinical Diagnosis. ScientificWorldJournal 2015;2015:657086 [PubMed]
- EL Andaloussi S. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov 2013;12:347-57. [Crossref] [PubMed]
- Colombo M, Raposo G, Théry C. Biogenesis, Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles. Annu Rev Cell Dev Biol 2014;30:255-89. [Crossref] [PubMed]
- Raposo G, Stoorvogel W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol 2013;200:373-83. [Crossref] [PubMed]
- Panagiotara A, Markou A, Lianidou ES, et al. Exosomes: A Cancer Theranostics Road Map. Public Health Genomics 2017;20:116-25. [Crossref] [PubMed]
- Yáñez-Mó M, Siljander PRM, Andreu Z, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles 2015;4:27066. [Crossref] [PubMed]
- Kalra H, Drummen GP, Mathivanan S. Focus on extracellular vesicles: Introducing the next small big thing. Int J Mol Sci 2016;17:170. [Crossref] [PubMed]
- Ludwig AK, Giebel B. Exosomes: Small vesicles participating in intercellular communication. Int J Biochem Cell Biol 2012;44:11-5. [Crossref] [PubMed]
- Valadi H, Ekström K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654-9. [Crossref] [PubMed]
- Keller S, Ridinger J, Rupp AK, et al. Body fluid derived exosomes as a novel template for clinical diagnostics. J Transl Med 2011;9:86. [Crossref] [PubMed]
- Li P, Kaslan M, Lee SH, et al. Progress in Exosome Isolation Techniques. Theranostics 2017;7:789-804. [Crossref] [PubMed]
- Enderle D, Spiel A, Coticchia CM, et al. Characterization of RNA from exosomes and other extracellular vesicles isolated by a novel spin column-based method. PLoS One 2015;10:e0136133 [Crossref] [PubMed]
- Fuhrmann G, Herrmann IK, Stevens MM. Cell-derived vesicles for drug therapy and diagnostics: Opportunities and challenges. Nano Today 2015;10:397-409. [Crossref] [PubMed]
- Whiteside TL. Tumor-Derived Exosomes and Their Role in Cancer Progression. Adv Clin Chem 2016;74:103-41. [Crossref] [PubMed]
- Braicu C, Tomuleasa C, Monroig P, et al. Exosomes as divine messengers: are they the Hermes of modern molecular oncology? Cell Death Differ 2015;22:34-45. [Crossref] [PubMed]
- Zhang X, Yuan X, Shi H, et al. Exosomes in cancer: small particle, big player. J Hematol Oncol 2015;8:83. [Crossref] [PubMed]
- Costa-Silva B, Aiello NM, Ocean AJ, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol 2015;17:816-26. [Crossref] [PubMed]
- Dutta S, Reamtong O, Panvongsa W, et al. Proteomics profiling of cholangiocarcinoma exosomes: A potential role of oncogenic protein transferring in cancer progression. Biochim Biophys Acta 2015;1852:1989-99. [Crossref] [PubMed]
- Haga H, Yan IK, Takahashi K, et al. Tumour cell-derived extracellular vesicles interact with mesenchymal stem cells to modulate the microenvironment and enhance cholangiocarcinoma growth. J Extracell Vesicles 2015;4:24900. [Crossref] [PubMed]
- Chen JH, Xiang JY, Ding GP, et al. Cholangiocarcinoma-derived exosomes inhibit the antitumor activity of cytokine-induced killer cells by down-regulating the secretion of tumor necrosis factor-α and perforin. J Zhejiang Univ Sci B 2016;17:537-44. [Crossref] [PubMed]
- Li L, Masica D, Ishida M, et al. Human bile contains MicroRNA-laden extracellular vesicles that can be used for cholangiocarcinoma diagnosis. Hepatology 2014;60:896-907. [Crossref] [PubMed]
- Letelier P, Riquelme I, Hernández AH, et al. Circulating MicroRNAs as biomarkers in biliary tract cancers. Int J Mol Sci 2016;17:E791 [Crossref] [PubMed]
- Masyuk AI, Huang BQ, Ward CJ, et al. Biliary exosomes influence cholangiocyte regulatory mechanisms and proliferation through interaction with primary cilia. Am J Physiol Gastrointest Liver Physiol 2010;299:G990-9. [Crossref] [PubMed]
- Tomiyama T, Yang GX, Zhao M, et al. The modulation of co-stimulatory molecules by circulating exosomes in primary biliary cirrhosis. Cell Mol Immunol 2017;14:276-84. [Crossref] [PubMed]
- Severino V, Dumonceau JM, Delhaye M, et al. Extracellular Vesicles in Bile as Markers of Malignant Biliary Stenoses. Gastroenterology 2017;153:495-504.e8. [Crossref] [PubMed]
- Wang Y, Wang G, Wang Z, et al. Chicken biliary exosomes enhance CD4 + T proliferation and inhibit ALV-J replication in liver. Biochem Cell Biol 2014;92:145-51. [Crossref] [PubMed]
- Allenson K, Castillo J, San Lucas FA, et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann Oncol 2017;28:741-7. [PubMed]
- Ahmad J, Singhal M, Amin S, et al. Bile salt stabilized vesicles (bilosomes): A novel nano-pharmaceutical design for oral delivery of proteins and peptides. Curr Pharm Des 2017;23:1575-88. [Crossref] [PubMed]
- Chaiyadet S, Sotillo J, Smout M, et al. Carcinogenic liver fluke secretes extracellular vesicles that promote cholangiocytes to adopt a tumorigenic phenotype. J Infect Dis 2015;212:1636-45. [Crossref] [PubMed]
- Hu G, Gong AY, Roth AL, et al. Release of Luminal Exosomes Contributes to TLR4-Mediated Epithelial Antimicrobial Defense. PLoS Pathog 2013;9:e1003261 [Crossref] [PubMed]
- Arbelaiz A, Azkargorta M, Krawczyk M, et al. Serum extracellular vesicles contain protein biomarkers for primary sclerosing cholangitis and cholangiocarcinoma. Hepatology 2017;66:1125-43. [Crossref] [PubMed]
- Victor DW, Sherman S, Karakan T, et al. Current endoscopic approach to indeterminate biliary strictures. World J Gastroenterol 2012;18:6197-205. [Crossref] [PubMed]
- Landskron G, De La Fuente M, Thuwajit P, et al. Chronic inflammation and cytokines in the tumor microenvironment. J Immunol Res 2014;2014:149185 [PubMed]


