
Transketolase contributes to hepatocellular carcinoma migration, invasion, angiogenesis, and tumorigenesis
Introduction
Transketolase (TKT) catalyzes the reversible transfer of two-carbon fragments between ketose and aldose phosphates by forming a homodimer (1). Crystal structure assays have suggested that TKT is divided into three domains: the N-terminal PP domain, the Pyr domain, and the C-terminal domain, which are interconnected by flexible linker regions; the active site is located in the interface between the PP and Pyr domains of both subunits (2). The pentose phosphate pathway (PPP) produces ribose-5-phosphate to regulate nucleotide synthesis, and also generates the antioxidant NADPH to promote cancer cell survival in oxidative stress. PPP is separated into the oxidative branch and the non-oxidative branch. The non-oxidative branch is mainly catalyzed by TKT; in many tumor cell lines, the non-oxidative branch synthesizes a major portion of the ribose in nucleic acids, suggesting that TKT regulates tumor progression (3,4). Meanwhile, phosphatidylinositol 3-kinase (PI3K)/AKT signaling is often deregulated in many tumors, and regulates tumor growth, proliferation, and survival (5,6). AKT phosphorylates TKT at Thr382, increasing its catalytic activity and the carbon flow through the non-oxidative PPP (7), indicating that TKT may play an important role in tumor development and progression. The role of TKT in tumor progression has not been well-studied, and there are few studies on TKT. The role of TKT in cervical cancer has been reported, where TKT is upregulated in metastatic peritoneal implants, and it promotes cervical cancer cell proliferation, but not motility and invasion. MicroRNA-497 increases the cisplatin chemosensitivity of cervical cancer cells by targeting TKT (8,9). However, the role of TKT in hepatocellular carcinoma (HCC) has not been studied.
HCC is one of the leading causes of cancer mortality worldwide, especially in Asia (10), and its recurrence rate after surgical resection, radiotherapy, and chemotherapy is high due to its higher metastatic potential. Screening new regulatory factors is vital for HCC therapy. Here, we studied the role of TKT in HCC progression. We found that TKT was upregulated in HCC cells and tissues, and patients with HCC with high TKT levels had poor outcomes. TKT also contributed to HCC migration, invasion, angiogenesis, and tumorigenesis. Moreover, there was lower TKT promoter methylation in HCC cells and tissues.
Methods
Cell culture and HCC specimens
HCC cells (SNU-475, Hep3B, Huh7, HepG2, Huh1, SK-Hep1) were purchased from American Type Culture Collection (ATCC) and cultured according to ATCC recommendations. The LO2 normal liver epithelial cell line was purchased from the Cell bank of the typical culture preservation Committee of Chinese academy of sciences, and cultured according to ATCC recommendations.
Three normal liver tissue specimens and nine HCC tissue specimens were collected from curative resection and were diagnosed histopathologically at the First Affiliated Hospital of Sun Yat-sen University. All samples were immediately frozen and stored in liquid nitrogen until used. A cohort of 88 HCC tissues was also collected from the First Affiliated Hospital of Sun Yat-sen University. Informed consent was obtained from all patients. The institutional research ethics committee approved the experimental protocols, the number of ethics approval was [2014]-12. The detailed clinicopathological characteristics were shown in Table S1.
Transfection
To overexpress TKT, we amplified the full-length TKT coding sequence using PCR and subcloned it into a pMSCV-retro-puro vector using the following forward and reverse primers: 5'-GAAGATCTATGGAGAGCTACCACAAGCC-3' and 5'-CGGAATTCCTAGGCCTTGGTGATGAGG-3', respectively. Empty vector was used as the negative control. The vectors were transfected into cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Puromycin (Sigma) was used to screen stable cell lines.
TKT small interference RNA (siTKT) modified with 2'-O-methyl and scramble control were purchased from Guangzhou RiboBio and transfected into cells using Lipofectamine 2000. The target sequence was 5'-TGCCATCATCTATAACAACAA- 3'.
Immunohistochemistry (IHC) assay
IHC assay was performed according to previously described methods (11,12) using anti-TKT antibody (sc-67120, Santa Cruz Biotechnology). The analysis was performed according to our previously described method (13). Briefly, the double-blind method was used to examine TKT levels in the HCC tissues. The proportion of tumor cells was scored as follows: 0 points, no positive cells; 1 point, 1–25% positive cells; 2 points, 26–50% positive cells; 3 points, 51–75% positive cells; 4 points, >75% positive cells. Protein expression intensity was scored as follows: 0 points (no staining), 1 point (weak staining, light yellow), 2 points (moderate staining, yellowish brown), or 3 points (strong staining, brown). The staining index was calculated as the sum of the staining intensity and the proportion of positive cell scores (0, 1, 2, 3, 4, 6, 8, 9 or 12 points). Cut-off values for TKT expression were chosen based on a measurement of heterogeneity using the log-rank test with respect to overall survival. The staining index which is less than 6 was considered to be low expression, and greater than or equal to 4 was considered to be high expression.
Reverse transcription-PCR (RT-PCR) and quantitative real-time PCR
Total RNA was isolated from cells or tissues using TRIzol (Life Technologies). Complementary DNA (cDNA) was synthesized using a HiScript II 1st Strand cDNA Synthesis Kit (Vazyme Biotech). Quantitative real-time PCR was performed using 7500 Fast Real-Time Sequence detection system software (Applied Biosystems). An AceQ qPCR SYBR Green Master Mix Kit (Vazyme Biotech) was used for analysis. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the internal control. Relative TKT expression was calculated using the comparative threshold cycle (2−ΔΔCt) method based on the threshold cycle (Ct) values. The forward and reverse primers used for the TKT quantitation were 5'-GCTGCTGAACCTGAGGAAGA-3' and 5'-TAGACTCGGTAGCTGGCCTT-3', respectively.
Western blotting
Total protein was extracted using radio immunoprecipitation assay buffer [50 mM Tris (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% NP-40, and 1% Triton X-100 supplemented with protease inhibitors (Roche)]. The primary antibodies used for western blotting were antibodies against TKT (sc-67120), matrix metalloproteinase (MMP)2 (sc-13594), MMP9 (sc-21733), vascular endothelial growth factor (VEGF) (sc-365578), and GAPDH (sc-365062) (all, Santa Cruz Biotechnology).
Wound healing assay
Cells were seeded in 6-well plates and cultured to confluence. Streaks were created in the cell monolayers using a 200-µL pipette tip, and then the wounding was observed and photographed at 0, 12, and 24 h.
Cell invasion assay
The cell invasion assay was performed according to our previously reported method (14). Briefly, cells were seeded on top of polycarbonate Transwell filters (8.0-µm pore size, Corning) and cultured with RPMI 1640 medium (Life Technologies) containing 5% fetal bovine serum (FBS, GIBCO); the bottom chamber was filled with RPMI 1640 medium containing 10% FBS. The cells were grown for 20 h, and migrated cells on the surface of the lower membrane were fixed with 4% paraformaldehyde, stained with hematoxylin for 15 min, and then photographed and counted.
Soft agar assay
The soft agar assay was performed according to our previously described methods (15).
Chicken chorioallantoic membrane (CAM) assay
The CAM assay was performed according to our previously described method (14).
Xenograft experiments
The animal studies were performed according to a protocol approved by the Sun Yat-sen University Institutional Animal Care and Use Committee. BALB/c nude mice (5–6 weeks old) were obtained from the Guangzhou University of Chinese Medicine Experimental Animal Center and were housed in facilities on a 12-h light/dark cycle, and five mice were randomly assigned to each treatment group. Cells (1×107) with TKT overexpression or knockdown were directly injected into the flanks of the mice. After 40 days, mice were sacrificed and the tumors were harvested and weighed.
Bisulfite genomic sequencing
Bisulfite treatment was carried out using an EpiTect Bisulfite Kit (Qiagen) according to the manufacturer’s protocol. The bisulfite sequencing PCR primers were designed using MethPrimer (http://www.urogene.org/methprimer/).
Statistical analysis
All statistical analyses were carried out using SPSS 20.0 (SPSS Inc.); a two-tailed paired Student t-test was used to compare two groups. The data are reported as the mean ± standard deviation (SD). Survival curves were plotted using the Kaplan–Meier method and were compared using the log-rank test. P<0.05 was considered statistically significant. Gene set enrichment analysis (GSEA) was performed using the GSEA application (http://software.broadinstitute.org/gsea/index.jsp).
Results
High TKT expression in HCC tissues correlated with poor survival
To determine the role of TKT in HCC progression, we first analyzed its expression. Analysis of the gene expression profiles of liver tumor tissues and normal liver tissues (GSE14520) revealed that TKT was significantly upregulated in liver tumor tissues as compared to the non-tumor tissues (Figure 1A) (16). The Cancer Genome Atlas (TCGA) data set also revealed that TKT was significantly upregulated in HCC tissues as compared to normal liver tissues (Figure 1B). GSEA suggested that TKT levels in HCC tissues were high as compared to the adjacent normal liver tissues (Figure 1C). These data suggest that TKT is upregulated in HCC tissues. To confirm this result, we determined TKT expression in HCC cells and tissues, where quantitative real-time PCR and western blotting revealed that TKT was upregulated in HCC cells as compared to the normal LO2 liver cells (Figure 1D). TKT was also upregulated in HCC tissues as compared to normal liver tissues (Figure 1E). These findings show TKT is upregulated in HCC cells and tissues.
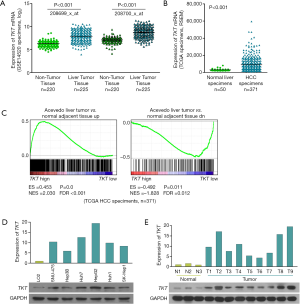
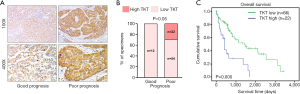
To determine the clinical significance of TKT expression, we examined TKT expression in 88 clinical specimens. We found 86 (97.7%) patients are TKT expression positive. Twenty-two (25.0%) patients had high TKT expression (Table S2). IHC suggested that patients with HCC with low TKT expression had good prognosis and that patients with HCC with high TKT expression had poor prognosis (Figure 2A). Statistical analysis also suggested that TKT expression was low in patients with HCC with good prognosis (12/12, 100%) and that TKT expression was high in patients with HCC with poor prognosis (22/76, 29%) (Figure 2B). Kaplan–Meier survival curves revealed that the overall survival of patients with HCC with high TKT expression was significantly shorter than that of patients with low TKT expression (Figure 2C). These findings suggest that high TKT expression in HCC tissues correlates with poor survival. We also analyzed TKT expression in malignant HCC and benign tumor in liver, like hepatic hemangioma, where GSEA determined that malignant HCC tissues had high TKT expression as compared to the benign tissues (Figure 3A). Altogether, TKT is upregulated in HCC cells and tissues, and patients with HCC with high TKT expression have poor clinical outcome, suggesting that TKT might be an oncogene for HCC progression.
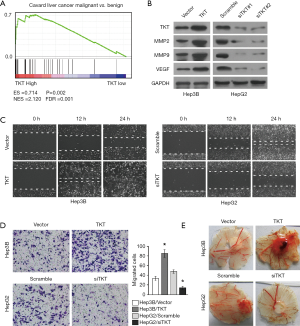
TKT promoted HCC migration, invasion, and angiogenesis
As tumor migration, invasion, metastasis, and angiogenesis are the main reasons for tumor relapse, we investigated the role of TKT in these aspects of HCC. We overexpressed TKT in Hep3B cells with low TKT expression, and downregulated TKT in HepG2 cells with high TKT expression to analyze the effect of TKT on HCC progression. MMP2, MMP9, and VEGF promote tumor migration, invasion, metastasis, and angiogenesis, and can function as tumor metastasis and angiogenesis markers (17-19). Western blotting revealed that TKT overexpression increased MMP2, MMP9, and VEGF expression, while TKT knockdown inhibited it (Figure 3B), suggesting that TKT regulates HCC migration, invasion, metastasis, and angiogenesis. We further confirmed this result by directly observing the phenotypic changes. The cell migration assay showed that TKT overexpression increased the migration ability of Hep3B cells, while TKT knockdown reduced the migration ability of HepG2 cells (Figure 3C). The cell invasion assay showed that TKT overexpression significantly promoted Hep3B cell invasive capability and that TKT knockdown significantly inhibited HepG2 cell invasive capability (Figure 3D). The CAM assay showed that TKT overexpression promoted angiogenesis, while TKT knockdown inhibited it (Figure 3E). These findings suggest that TKT contributes to HCC migration, invasion, and angiogenesis, and might be the reason for HCC relapse.
TKT promoted HCC tumorigenesis
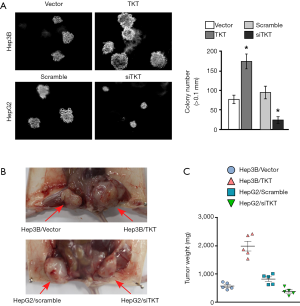
The soft agar assay showed that TKT overexpression significantly increased tumorigenesis ability in vitro and that TKT knockdown significantly inhibited it (Figure 4A). The xenograft tumor model showed that TKT overexpression increased tumorigenesis in vivo and that TKT knockdown reduced it (Figure 4B,C). Altogether, TKT contributed to tumorigenesis and was an oncogene for hepatocellular tumorigenesis.
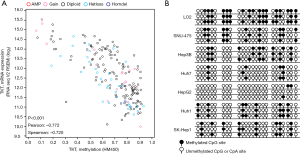
As promoter DNA methylation is a marker of gene silencing (20), we analyzed the correlation between TKT expression and the methylation level of the TKT promoter in HCC tissues, and found a negative correlation between TKT expression and TKT promoter methylation levels, suggesting that TKT promoter methylation caused the TKT upregulation in HCC tissues (Figure 5A). Bisulfite genomic sequencing also revealed lower TKT promoter methylation in HCC cells as compared to the normal LO2 liver cells (Figure 5B). This finding suggests that TKT is upregulated in HCC cells and tissues due to lower TKT promoter methylation.
Discussion
In the present study, we found that TKT was upregulated in HCC tissues and cells. Patients with HCC with poor prognosis had high TKT expression, and the opposite was true for patients with low TKT expression. TKT overexpression promoted HCC cell migration, invasion, angiogenesis, and tumorigenesis, while TKT knockdown reduced these effects. We also found lower TKT promoter methylation in HCC tissues and cells.
Our study suggests that TKT is an oncogene for HCC progression, promoting HCC migration, invasion, angiogenesis, and tumorigenesis; however, its regulatory mechanism is not understood. c-Myc is a well-known oncogene; many studies have shown that its deregulation is important for HCC initiation and progression and that it promotes hepatocarcinogenesis, proliferation, growth, invasion, and migration (21). Proteomics assays have suggested that TKT interacts with c-Myc (22); therefore, TKT might promote HCC progression by increasing c-Myc activity.
TKT also interacts with H2A histone family member X (H2AX) in HCC cells (23); H2AX plays a central role in cellular responses to DNA damage and DNA damage repair. Phosphorylated histone H2AX (γ-H2AX) regulates DNA repair and is a DNA damage marker. H2AX and its interacting proteins play synergistic roles in tumor progression; in different tumors, H2AX interacts with different proteins (24,25). We found that TKT was upregulated in HCC cells and tissues, and contributed to HCC progression, and inferred that TKT might regulate DNA damage repair. However, the detailed mechanisms require further study. Recently, Xu et al. found that the nuclear factor erythroid 2–like 2/Kelch-like ECH-associated protein 1/BTB domain and CNC homolog 1 (NRF2/KEAP1/BACH1) oxidative stress sensor pathway regulates TKT expression in HCC, where TKT inhibition increased the sensitivity to sorafenib (26).
In summary, we found that TKT is an oncogene for hepatocellular tumorigenesis and that it promotes HCC migration, invasion, angiogenesis, and tumorigenesis. However, its regulatory mechanism is not understood.
Full table

Full table
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (No. 30600156, and 81171442, and 81602701), the Natural Science Foundation of Guangdong Province, China (No. 2014A030313090, 2014A030313190, and 2017A030313547), the Science and Technology Projects Foundation of Guangdong Province, China (No. 2015A070710006 and 2016A020215053), and the Science and Technology Projects Foundation of Guangzhou City, China (No. 201507020037 and 201607010260).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.10.26). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The institutional research ethics committee approved the experimental protocols and the number of ethics approval was [2014]-12. Informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kochetov GA. Functional flexibility of the transketolase molecule. Biochemistry (Mosc) 2001;66:1077-85. [Crossref] [PubMed]
- Mitschke L, Parthier C, Schröder-Tittmann K, et al. The crystal structure of human transketolase and new insights into its mode of action. J Biol Chem 2010;285:31559-70. [Crossref] [PubMed]
- Pácal L, Tomandl J, Svojanovsky J, et al. Role of thiamine status and genetic variability in transketolase and other pentose phosphate cycle enzymes in the progression of diabetic nephropathy. Nephrol Dial Transplant 2011;26:1229-36. [Crossref] [PubMed]
- Vizán P, Alcarraz-Vizán G, Díaz-Moralli S, et al. Modulation of pentose phosphate pathway during cell cycle progression in human colon adenocarcinoma cell line HT29. Int J Cancer 2009;124:2789-96. [Crossref] [PubMed]
- Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer 2002;2:489-501. [Crossref] [PubMed]
- Luo J, Manning BD, Cantley LC. Targeting the PI3K-Akt pathway in human cancer: rationale and promise. Cancer Cell 2003;4:257-62. [Crossref] [PubMed]
- Saha A, Connelly S, Jiang J, et al. Akt phosphorylation and regulation of transketolase is a nodal point for amino acid control of purine synthesis. Mol Cell 2014;55:264-76. [Crossref] [PubMed]
- Ricciardelli C, Lokman NA, Cheruvu S, et al. Transketolase is upregulated in metastatic peritoneal implants and promotes ovarian cancer cell proliferation. Clin Exp Metastasis 2015;32:441-55. [Crossref] [PubMed]
- Yang H, Wu XL, Wu KH, et al. MicroRNA-497 regulates cisplatin chemosensitivity of cervical cancer by targeting transketolase. Am J Cancer Res 2016;6:2690-9. [PubMed]
- Tan G, Wu L, Tan J, et al. MiR-1180 promotes apoptotic resistance to human hepatocellular carcinoma via activation of NF-kappaB signaling pathway. Sci Rep 2016;6:22328. [Crossref] [PubMed]
- Cheang TY, Zhou HY, Chen W, et al. C14orf166 overexpression correlates with tumor progression and poor prognosis of breast cancer. J Transl Med 2016;14:54. [Crossref] [PubMed]
- Liu Q, Tu K, Zhang H, et al. TPX2 as a novel prognostic biomarker for hepatocellular carcinoma. Hepatol Res 2015;45:906-18. [Crossref] [PubMed]
- Fu B, Meng W, Zhao H, et al. GRAM domain-containing protein 1A (GRAMD1A) promotes the expansion of hepatocellular carcinoma stem cell and hepatocellular carcinoma growth through STAT5. Sci Rep 2016;6:31963. [Crossref] [PubMed]
- Xing S, Zhang B, Hua R, et al. URG4/URGCP enhances the angiogenic capacity of human hepatocellular carcinoma cells in vitro via activation of the NF-kappaB signaling pathway. BMC Cancer 2015;15:368. [Crossref] [PubMed]
- Li H, Zheng D, Zhang B, et al. Mir-208 promotes cell proliferation by repressing SOX6 expression in human esophageal squamous cell carcinoma. J Transl Med 2014;12:196. [Crossref] [PubMed]
- Roessler S, Jia HL, Budhu A, et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res 2010;70:10202-12. [Crossref] [PubMed]
- Tang Y, Lv P, Sun Z, et al. 14-3-3β Promotes migration and invasion of human hepatocellular carcinoma cells by modulating expression of MMP2 and MMP9 through PI3K/Akt/NF-κB Pathway. PLoS One 2016;11:e0146070 [Crossref] [PubMed]
- Hu J, Ni S, Cao Y, et al. The angiogenic effect of microRNA-21 targeting TIMP3 through the regulation of MMP2 and MMP9. PLoS One 2016;11:e0149537 [Crossref] [PubMed]
- Sitohy B, Nagy JA, Dvorak HF. Anti-VEGF/VEGFR therapy for cancer: reassessing the target. Cancer Res 2012;72:1909-14. [Crossref] [PubMed]
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet 2008;9:465-76. [Crossref] [PubMed]
- Zimonjic DB, Popescu NC. Role of DLC1 tumor suppressor gene and MYC oncogene in pathogenesis of human hepatocellular carcinoma: potential prospects for combined targeted therapeutics Int J Oncol 2012;41:393-406. (review). [Crossref] [PubMed]
- Agrawal P, Yu K, Salomon AR, et al. Proteomic profiling of Myc-associated proteins. Cell Cycle 2010;9:4908-21. [Crossref] [PubMed]
- Yang X, Zou P, Yao J, et al. Proteomic dissection of cell type-specific H2AX-interacting protein complex associated with hepatocellular carcinoma. J Proteome Res 2010;9:1402-15. [Crossref] [PubMed]
- Matsuda Y, Wakai T, Kubota M, et al. DNA damage sensor gamma -H2AX is increased in preneoplastic lesions of hepatocellular carcinoma. ScientificWorldJournal 2013;2013:597095 [PubMed]
- Raymond AA, Benhamouche S, Neaud V, et al. Reptin regulates DNA double strand breaks repair in human hepatocellular carcinoma. PLoS One 2015;10:e0123333 [Crossref] [PubMed]
- Xu IM, Lai RK, Lin SH, et al. Transketolase counteracts oxidative stress to drive cancer development. Proc Natl Acad Sci U S A 2016;113:E725-34. [Crossref] [PubMed]


