
Study the effect of CTSD interaction proteins in invasion and metastasis of nasopharyngeal carcinoma
Introduction
Invasion and metastasis were considered to be the most important prognostic factors and the leading causes of death in cancer patients. Nasopharyngeal carcinoma (NPC) is a malignant tumor of head and neck, which arises in surface epithelium of the posterior wall of the nasopharynx. Concomitant chemoradiotherapy followed by adjuvant chemotherapy was the preferred therapeutic option (1). Epidemiologic studies highlight a connection between Epstein-Barr virus (EBV) and NPC (2,3). Because of its deep location and high metastatic potential, the prognosis of NPC patient is poor (4). To reduce the risk of distant metastasis of NPC and increase survival rate of NPC patients, elucidation of the molecular mechanism of NPC has been in urgent need.
Tumor invasion and metastasis is a multi-step process. Metastasis is an inherent property of cancer cells, which is also modulated by the components of tumor microenvironment, such as extracellular matrix (ECM). Tumor cells express a variety of proteases from matrix metalloproteinases (MMPs), cathepsins, which could surmount ECM barriers (5).The interaction of tumor cells with local stroma plays a critical role during metastatic dissemination (6). Several components of ECM, such as fibronectin, proteoglycan, and collagen, are substrates of lysosomal aspartic protease (7-9). Lysosomes maintain cellular metabolism by degrading unneeded extracellular and intracellular substances (10). It has been reported that tumor invasion and metastasis are associated with altered lysosomal trafficking and increased expression of lysosomal proteases termed cathepsins (11). Cathepsin D is a soluble aspartic lysosomal protease, which extensively presents in mammalian cells. It is synthesized as an inactive pre-proenzyme, of which the amino terminal peptides get proteolytic cleavage within the acidic environment of endosomal and lysosomal compartments (12). Cathepsin D is released from lysosomes into cytosol to be functional. With its enzymatic property, cathepsin D regulates a number of important cellular processes including cell growth, proliferation, motility, antigen processing, and so on.
So far, few attempts have been made for proteomic-wide screening for proteins interacting with cathepsin D. In our previous study, comparative proteomic analysis was performed to identify differential expression proteins between the NPC and normal nasopharyngeal epithelial tissue (NNET). In the meanwhile, considerable evidences suggest that cathepsin D may be involved in the invasion and metastasis of NPC. Here we employed co-immunoprecipitation (co-IP)/mass spectrometry (MS)-based proteomics, followed by stringent bioinformatic analysis and validation strategy to elucidate the mechanisms how cathepsin D promotes invasion and metastasis. We identified 141 cathepsin D interacting proteins. Most of the identified proteins have been reported to be associated with tumor invasion and metastasis. Among these, epidermal growth factor receptor (EGFR), HSP90A were confirmed by co-IP followed by immunoblotting. These two proteins have been known as tumor-associated proteins and have been implicated in important cellular processes, such as cell growth, apoptosis and malignant transformation etc. In the present study, we found that cathepsin D/EGFR/HSP90A form a complex and cathepsin D may mediate NPC cell invasion and migration by regulating the function of EGFR and HSP90A.
Methods
Reagents
PVDF membrane and ZipTip C18 columns were obtained from Millipore (Boston, MA, USA). The anti-cathepsin D antibody, anti-EGFR antibody, and anti-HSP90A antibody were purchased from Santa Cruz Biotechnology, Inc (Santa Cruz, CA, USA). IgY was purchased from Jackson ImmunoResearch Laboratories (West Grove, PA, USA). Protein-G sepharose beads were purchased from GE Healthcare Life Sciences (Marlborough, MA, USA). Trypsin and ECL reagent were purchased from Amersham Biosciences (Stockholm, Sweden). All buffers were prepared with Milli-Q water.
Cell and tissue samples
NPC 5-8F cell line with high metastatic potential and 6-10B cell line without metastatic potential were kindly provided by the key laboratory of cancer proteomics of Chinese ministry of health, Xiangya Hospital, cultured in RPMI-1640 (Sigma, St. Louis, MO, USA) medium supplemented with 10% fetal bovine serum (Hyclone), 100 IU/mL penicillin and 100 µg/mL streptomycin. The culture flasks were incubated at a humidified atmosphere of 5% CO2 at 37 °C. Cells were harvested when 80% of confluency was attained. The cell pellets were suspended in lysis buffer containing 20 mmol/L Tris, Ph 7.4, 100 mmol/L NaCl2, 1% NP-40, 0.5 mmol/L EDTA, 0.5 mmol/L Na3VO4, and 0.5 mmol/L PMSF. The lysates were then centrifuged at 13,000 rpm for 20 min. The supernatant was collected and the protein concentrations were quantified using Bradford assay. Fifty-eight case formalin-fixed and paraffin-embedded poorly differentiated squamous cell carcinomas tissues were obtained from the Department of Pathology, Xiangya Hospital of Central South University, China, at the time of diagnosis before any therapy.
Co-IP
5-8F cell lysates containing 1.5 mg of the protein were first precleared using pre-immune serum and 100 µL of protein G-Sepharose beads (GE HealthCare Life Sciences, Marlborough, MA, USA) at 4 °C shaking 2 hours, used as control for nonspecific IP. The lysates with beads were then centrifuged for 5 min at 9,000 rpm, the supernatant were removed into a fresh tube. The testing samples were immunoprecipitated using 10 µg of anti-cathepsin D antibody, incubated 3 hours, then 100 µL of protein G-Sepharose beads were added for 1 hour, 4 °C. The pelleted beads were washed three times with TBS-T buffer after centrifuge, and eluted with 0.1 M glycine from pH 3.0 adjusted to pH 7.5 with Tris buffer. Immunoprecipitates were washed 3 times, and then eluted with SDS sample buffer containing 2-mercaptoethanol at 100 °C for 3 min. IgY was used instead of cathepsin D antibody for control group.
SDS-PAGE
The proteins were then separated by 12.5% SDS-PAGE, and transferred onto PVDF filters. The same procedure was repeated with IPs from cells lysates as a control. After electrophoresis, the protein bands in the gel was visualized by Coomassie blue R-250 (Amersham Biosciences, Stockholm, Sweden) and then destained with three changes of destain buffer, 1 hour each. Other standard chemical reagents required were purchased from Sigma. Stained gels were scanned on an Image-Scanner (Amersham Biosciences, Stockholm, Sweden). Protein bands detected in cathepsin D immunoprecipitation sample but not present in the controls were excised from the gel, and diluted into a 1.5 mL sterile microcentrifuge tube.
MS analysis and database searching
The differential protein bands were dissolve in digestion buffer, digested with trypsin, then destained with 50% acetonitrile (ACN)/100 Mm, ammonium bicarbonate (NH4HCO3; 100 µL) for 30 min with constant mixing. After removal of the solution, the gel pieces were dehydrated with CAN (100 µL) for 15 min at room temperature (RT) and dried in a spread vac concentrator. The dried gel-pieces were incubated in 10 µL digestion solution consisted of 40 Mm NH4HCO3 in 9% ACN solution and 20 g/mL proteomics grade trypsin for 10–12 hours at 37 °C for 14–16 hours. The digests were extracted and purified with Millipore ZipTic C18 column. Samples were loaded, trapped and washed at a flow rate of 30 µL/min with 99% solvent A (0.1% FA)/1% solvent B (CAN containing 0.1 FA), C 0.1% FA 5% ACN. Peptides were eluted with a gradient of 1% to 50% B for 50 min, 95% B for 10 min, and then 99% A for 10 min before loaded on a pre-column (320 µm × 5 mm, 5 µm C18 silica beads, waters) at 30 µL/min. Then samples were loaded onto a precolumn for concentrations and fast desalting through a Waters CapLC autosampler, and then eluted to the reversed phase column (75 µm × 150 mm, 5 µm, 100 A, LC packing) at a flow rate of 200 nL/min after flow splitting for separation. MS/MS spectra were performed in data-depended mode in which up to 4 precursor ions above an intensity threshold of 7 counts/s (cps) were selected for MS/MS analysis from each survey ‘scan’. The nanospray parameters were 3,000 V for capillary voltage, 45 V for cone voltage, 80 °C for source temperature, and 15 psi collision gas back pressure. Proteins were analyzed by liquid. Chromatography-nanoelectrospray-tandem mass spectrometry (LC-ESI-MS/MS) uses a Waters electrospray ionization quadrupole time of flight 2 (ESI-Q-TOF2) system. The tandem MS data was processed and transformed to the PKL file format using Masslynx V 4.0 software (Micromass, Cary, USA) and then imported into an local Macot search engine (Matrix Science Ltd, London, UK) for protein identify searching. MS was searched against the NCBI nonredundant protein database. The search was restricted to ‘homo sapiens (human)’ as taxonomy. The other main search parameters were as follows: enzyme, trypsin; fixed modification, carbamidomethylcysteine; variable modifications, oxidation (M); fragment mass tolerance, 0.3 Da; peptide charge state, 1+, 2+, and 3+, and maximum missed cleavages, 1. Proteins whose scores were greater than 31 were considered significant (P<0.05).
Bioinformatics analysis
For functional interpretation of the resultant protein list from MS, the protein data were uploaded into the DAVID software. Gene ontology (GO), signaling pathways analysis and genes functional classifications were conducted. The protein-protein interaction networks were mapped by the STRING, a search tool for the Retrieval of interacting Genes/Proteins system (http://string-db.org/). Molecule functions classification and cluster analysis were performed through GO and cluster program of DAVID (http://david.abcc.ncifcrf.gov/). The Mascot scores were obtained from Mascot (Matrix Science, Torrance, London, UK). The protein coverage was reported. Biocarta and KEGG signaling pathways analysis were performed. Protein interaction network showed that EGFR, HSP90A and cathepsin D may constitute interactome, which provide new clues for the mechanism study of cathepsin D in invasion and metastasis of NPC.
Western blotting verification for bioinformatics analysis
To confirm cathepsin D associated protein EGFR and HSP90A, which were isolated from SDS-PAGE gel and identified by MS proteomic approach, western blotting was used for verification. Protein lysates were separated by 12.5% SDS-PAGE, and transferred onto PVDF filters, blocked with 5% non-fat milk in Tris-buffered saline, and followed by overnight incubation at 4 °C with antibodies against cathepsin D (Santa Cruz Biotechnology, Inc., USA), EGFR, HSP90A, then followed by 1 hour incubation with their corresponding secondary antibodies conjugated with horseradish peroxidase. ECL was used as detection reagents (Amersham Biosciences).
Reverse co-IP and western blotting confirmation for protein-protein interaction
The cell lysates were divided into four groups, co-IP with antibodies targeting cathepsin D, EGFR, HSP90A respectively, and IgY was used as negative control. The proteins were then electrotransferred and probed with monoclonal antibody against cathepsin D, EGFR and HSP90A.
Immunohistochemistry (IHC)
Tissue sections were deparaffinized with 100% xylene and rehydrated in descending percentage of ethanol series according to standard protocol. Heat-induced antigen retrieval was performed in 10 Mm citrate buffer for 15 min at 100 °C. Samples were incubated in endogenous peroxidase activity and nonspecific antigen solution, followed by incubation with anti-cathepsin D antibody (1:150), anti-EGFR or anti-HSP90A at 4 °C overnight, and then incubated with avidin-biotin peroxidase complex (DAKO, Carpinteria, CA, USA). Sections were visualized with DAB and counterstained with Harris’ modified hematoxylin, analyzed using light microscopy. The stained tissue sections were blindly reviewed and scored by two pathologists to the clinical parameters. For the negative control, the primary antibody was replaced by phosphate buffered saline (PBS). And 10 high-power fields were chosen randomly. The extent of the staining defined as the percentage of positive staining areas in relation to the whole section area.
The intensity of staining was graded on the following scale: 0, no staining; 1+, mild staining; 2+, moderate staining; 3+, intense staining. The area of staining was evaluated as follows: <5% stained positive, no staining of cells in any microscopic fields; 1+, between 6% and 30% stained positive; 2+, between 30% and 60% stained positive; 3+, >60% stained positive. A combined staining score <2 was considered to be a negative staining (low staining); a score between 3 and 4 was considered to be a moderate staining; whereas a score between 5 and 6 was considered to be a strong staining.
Transwell invasion assay
To study whether cathepsin D, EGFR, HSP90A affect invasive ability of NPC cells, high metastatic 5-8F cells with cathepsin D high expression were stably transfected with cathepsin D siRNA in pSilencer vector and control pSilencer-Scr, cell lines 5-8F-siRNA(+) and 5-8F-siRNA(-) were established respectively. non-metastatic 6-10B cells with cathepsin D low expression were stably transfected with cathepsin D-expressing vector pcDNA3-HA-cathepsin D and control vector pcDNA3, and the cell lines 6-10B-cathepsin D(+) and 6-10B-cathepsin D(-) were established, respectively. Cell invasion was estimated using 24-well transwell chambers (Costar, Cambridge, MA, USA): the upper and lower culture compartments of each well were separated by polycarbonate membranes (8-µm pore size). Briefly, for invasion assay, the membrane was precoated with 100 µg/cm2 membrane matrix; 0.5 mL of serum free RPMI-1640 medium was added to the well for 2 hours. In order to assess the ability of the cells to penetrate the precoated polycarbonate membrane, 1.25×104 cells were seeded in the upper chamber with 0.5 mL medium containing 1% FBS. The lower chamber contained 0.75 mL medium containing 10% FBS. After 24 hours incubation at 37 °C in a 5% CO2 incubator, the cells that had migrated to the underside were stained with a Diff-Quik stain kit (Dade Behring, Newark, DE, USA). Cells that reached the underside of the filter were counted in five random fields under a microscope (original magnification, ×200). Invasive ability was defined as the average cell numbers that penetrated the matrix-coated membrane per field. Three independent experiments were repeated.
Statistical analysis
All data were statistical analyzed for using SPSS version 17.0 statistical software. t-test was employed to analyze the differences in transwell cell invasiveness among different groups. P<0.05 was considered to be statistically significance.
Results
One hundred and forty-one cathepsin D-associated proteins are identified by co-IP followed by MS
To gain deep insight into the molecular mechanisms by which cathepsin D enhances cancer cell invasion, we examined the protein fractions isolated from high metastasis NPC 5-8F cell. 5-8F NPC cell lysate was collected, and incubated with anti-cathepsin D antibody and protein G-Sepharose beads for co-IP, which was then followed by SDS-PAGE western blotting analysis (Figure 1).
The gels were stained with Deep Purple and specific bands were excised and digested with trypsin, analyzed using ESI-Q-TOF-MS. One hundred and forty-one cathepsin D associated proteins were identified by subtraction of proteins present in the negative control (Table S1).
Validation of the cathepsin D interaction proteins by western blot
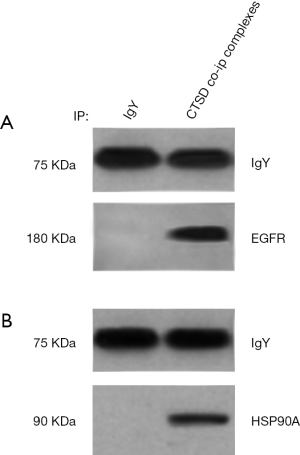
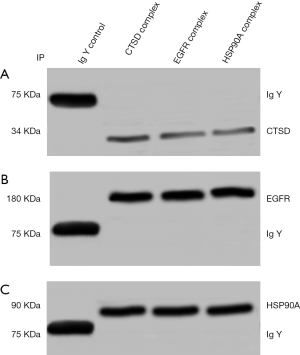
To validate the protein list generated by MS analysis, we checked two proteins from the list: EGFR and Hsp90A by co-IP followed by western blotting. Equal amounts of NPC 5-8F cell lysates was subjected for co-IP with antibody against cathepsin D followed by western blotting. As shown in Figure 2, consistent with the results of bioinformatics analysis, EGFR and HSP90A were able to be detected in western blot experiment by their corresponding antibodies, from the protein complex pulled down by cathepsin D antibody. No detection was observed in IgY negative control. These results suggest that the MS based identification of cathepsin D interaction proteins are reliable (Figure 2).
Bioinformatics strategy for interpretation of the proteomic data
The identified proteins were grouped according to DAVID (http://david.abcc.ncifcrf.gov) software guideline. GO-biological process (BP), GO-molecular function (MF) analysis, clustered these proteins into 12 functional categories. Gene Functional Classification showed that 70 out of 141 cathepsin D interaction proteins were divided into 12 classes. The functions of these proteins mainly included: transmembrane transport, cytoskeleton, oxidative phosphorylation, protein synthesis, cell apoptosis, signal transfer, oxidoreduction, molecular chaperone, glycometabolism, etc. The other 71 cathepsin D interaction proteins were not clustered by the software. GO-BP analysis showed that the BPs of 141 cathepsin D interaction proteins were mainly involved in stress reaction, negative control, metabolism, transport, localization, positioning, etc. GO-MF analysis showed that the MFs of cathepsin D interaction proteins were involved in protein binding, catalytic activity, purine nucleotide binding, cytoskeleton, oxidation reduction, molecular structure, etc. GO-CC analysis showed that the cathepsin D interaction proteins were localized in membrane, organelle, vesicle, polymer composites, etc. (data not shown). These suggest cathepsin D is involved in a number of cell signaling and metabolic processes in NPC metastasis as well as its known enzymatic activity.
Protein-protein interaction analysis
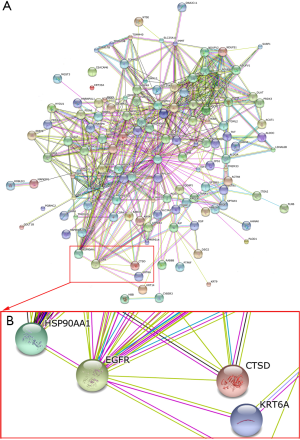
To map the correlation of these cathepsin D association proteins using string software (http://string-db.org), as shown in Figure 3, cathepsin D interacts with EGFR, HSP90A, suggesting that cathepsin D/EGFR/HSP90A may play an important role in NPC invasion and metastasis.
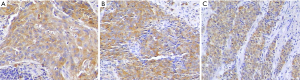
Correlation analysis of cathepsin D/EGFR/ HSP90A expression in NPC tissues visualized by IHC
IHC was performed to detect the expression of cathepsin D, EGFR, HSP90A in 58 cases of NPC (Figure 4). The correlation among the expression of cathepsin D, EGFR and HSP90A in NPC were analyzed by Spearman. When HSP90A was used as a control variable, EGFR expression was positive correlated with cathepsin D expression (Table 1, r=0.418, P=0.001); when EGFR was used as a control variable, HSP90A expression was positive correlated with cathepsin D expression (Table 1, r=0.373, P=0.004); however, when cathepsin D was used as a control variable, HSP90A expression was not correlated with EGFR expression (data not shown). These results indicated that the expression of cathepsin D was positive correlated with that of EGFR and HSP90A in NPC tissues.

Full table
Cathepsin D /EGFR/ HSP90A formed complexes in NPC cells
In order to further verify the interactions among cathepsin D/EGFR/HSP90A, co-IP combined with western blot analysis was used. EGFR and HSP90A band were detected by anti-EGFR, anti-HSP90A antibody respectively in cathepsin D co-IP complexes pulled down from NPC 5-8F cell lysate. As shown in Figure 5, the result suggested cathepsin D/EGFR/HSP90A formed complex in the NPC cells.
Cathepsin D modulated the expression of EGFR and HSP90A and enhanced in vitro invasiveness of NPC cells
The expressions of cathepsin D, EGFR and HSP90A in these reconstructed cells were determined by western blot. As shown in Figure 6A,B, compared to the control, introduction of cathepsin D siRNA into 5-8F cells significantly decreased the expressions of EGFR and HSP90A; while overexpression of cathepsin D in 6-10B cells significantly increased EGFR and HSP90A expressions. The results showed that cathepsin D expression was positive associated with the expression of EGFR and HSP90A.
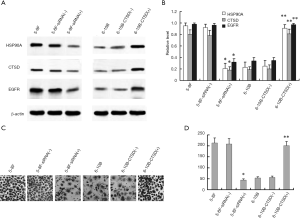
To evaluate the effect of cathepsin D modulation on the invasion of NPC cells, an in vitro invasion assay was used. As shown in Figure 6C,D, invasive cells in 5-8F-siRNA(+) were about 3.8-fold less than in 5-8F-siRNA(−) (P<0.01), and invasive cells in 6-10B-cathepsin D(+) were about 3.5-fold higher than in 6-10B-cathepsin D(−) (P<0.01). The results suggested that the expression levels of cathepsin D were associated with the in vitro invasive ability of NPC cells. Taken together, the results suggested that cathepsin D could promote invasion of NPC cells in vitro and possibly through cathepsin D/EGFR/HSP90A interactions.
Discussion
Proteins execute various functions by interacting with different sets of protein partners. By identifying the interacting partners, novel functions of poorly characterized proteins could be explored.
Cathepsins are lysosomal hydrolytic enzymes with ubiquitous presence in virtually all animal cells. Cathepsins harbor three subgroups according to the amino acid constituent at its active site: cysteine (B, C, H, F, K, L, O, S, V and W), aspartic (D and E), and serine (G) cathepsins (13). Cathepsin D is one of major components of lysosomes (14). Growing evidences show that they are involved in the destruction of tissue barriers during the invasion process of malignant tumors. Myb-binding was found in cathepsin D complex. In the control of specific proteases strongly increased the expression of cathepsin D, regulate the matrix-dependent invasion of breast cancer cells (15). Myb-binding protein 1A was found to interact with cathepsin D, providing further support that cathepsin D functions as a regulatory molecule during NPC migration and invasiveness. Antigen process in employing cathepsins B, D, and/or E for digesting protein Ag, is capable of processing complex protein Ag into antigenic peptides (16). Overexpression of AGR2 induced upregulation of CTSB and cathepsin D. AGR2-induced invasion was mediated through the activity of these proteases rather than by increased cell motility (17). It has been suggested that Rab27a affects the invasive and metastatic potential of breast cancer cells by modulating the secretion of cathepsin D as well as IGF-IL (18). Moreover, it has been reported that RNAi-Rab 27A inhibited both the lysosomal exocytosis of cathepsin D and cathepsin D enzyme activity, consequently inhibiting glioma cell migration (19). These observations imply that it is the protein-binding activity of cathepsin D that may be involved in stimulation of the tumor cells.
Although it seems that cathepsin D plays a role in protein degradation in a non-specific manner in the acidic environment of lysosomes, cathepsin D interacts with partner molecules and turns on signaling pathway in a specific manner. For example, Benes et al has shown that apoptosis is regulated by catalytically inactive mutants of cathepsin D which suggests that the enzymatic activity of cathepsin D influences cell apoptosis signaling (7). From protein-protein interaction databases, over 50 interaction partners for cathepsin D from different cellular compartments were indicated, suggesting the multiple-function role of cathepsin D (12). Protein-protein interaction network showed that PHB (Prohibitin) and cathepsin D networks have three interconnecting proteins: solute carrier family 2, facilitated glucose transport member 4 (glucose transporter 4, insulin-responsive) and APOA1 binging protein. Cathepsin D interacts with C-Myb to promote MDA-MB-231 cells migration and invasion (15). Cathepsin D expression was reduced by NESG1. NESG1 inhibited NPC invasion partly by downregulating cathepsin D. Cathepsin D was identified to be regulated by NESG1 (19) as NESG1 potential tumor suppressor in NPC cell. Cathepsin D mediates apoptosis induced by various agents, for example, interferon-r, Fas/Apo and tumor necrosis factor-α (20).
Our previous studies using LCM combined with MS have demonstrated that cathepsin D are differential proteins between NPC and NNETs (21). In subsequent experiments, IHC was performed and it was found that significant down-regulation of cathepsin D was observed in NPC but not in NNET. Up-regulation of cathepsin D was observed in lymph node metastasis versus primary NPC. Down-regulation of cathepsin D by siRNA significantly decreased in vitro invasive ability of 5-8F cells. Cathepsin D up-regulation was significantly correlated with advanced clinical stage, recurrence, lymph node and distant metastasis, indicating that it has a role in promoting NPC metastasis (22). However, little is known regarding the mechanism how cathepsin D promotes NPC metastasis.
In this study, we used co-IP combined with MS and identified 141 cathepsin D interaction proteins in NPC cell. Two candidates were selected for further validation by western. We confirmed that EGFR and HSP90A did interact with cathepsin D, demonstrating the reliability of the MS as the method in identifying protein complex. Biological interpretation of large genetic information derived from high-throughput genomic or proteomic studies can be challenging. More effort in exploration and data mining through high-throughput data is in need. Efficient tools are needed to gather, display and facilitate analysis of large data. Due to large number of components and complex BPs, visual tools are useful in providing an overview of networks. Through proteomics analysis, we have carried out a Gene Functional Classification screen of the proteins that interact with cathepsin D, 70 cathepsin D interaction proteins in 141 cathepsin D interaction proteins were grouped into 12 sets. The functions of these proteins mainly involve: transmembrane transport, cytoskeleton, oxidative phosphorylation, protein synthesis, cell apoptosis, signal transfer, oxidoreduction, molecular chaperone, glycometabolism, etc. We demonstrate that cathepsin D facilitates migration/invasion by interacting with EGFR, HSP90A.
Heat-shock-proteins (HSP) are ubiquitous and highly conserved in prokaryotes and eukaryote organisms. It has multiple functions involved in cell cycling, cell growth, DNA transcription and apoptosis (23-26). HSP expression is induced by heat and stresses including radiation and cytotoxic chemotherapy exposure. Based upon their molecular weight, amino acid sequence homology, and function, mammalian HSPs have been classified into five major families HSP100, HSP90, HSP70, HSP60, and the small Hsps. Among these, molecular chaperone HSP90 (90 kDa heat-shock protein) is the most abundant cytosolic HSP. The overexpression of HSP has been documented in hematologic malignancies (27) and solid tumors (28). It may contribute to drug resistance and it is related to poor prognosis (29). HSP90 has two isoforms, HSP90α (HSP90A) and HSP90β (constitutively active form). HSP90A were found to interact with cathepsin D in our study. Since HSP90A plays an important role in cell proliferation, we chose HSP90A for further test and verification.
The EGFR, which was identified in the complex with cathepsin D, is a member of the ErbB family of receptor tyrosine kinases. EGFR is expressed in tissues of epithelial, mesenchymal and neuronal origin (30). There are six known direct binding ligands for EGFR, EGF, transforming growth factor, amphiregulin, betacellulin, epiregulin, and heparin-binding EGF (31). This protein plays an important role in physiological processes, including differentiation, proliferation, and development (32). EGFR also functions in various pathological processes essential for cancer development, including cell division, angiogenesis, migration, and inhibition of apoptosis (33). EGFR is able to transduce extra-cellular mitogenic signals, such as EGF and transforming growth factor-alpha (TGF-α), by activating downstream signaling cascades. The downstream signalings involve components of phospholipase C-c, Ras, and phosphatidylinositol-3 kinase (PI-3K) (34). The pathway that leads to the suppression of apoptosis through PI3K and Akt activation, favors the development and progression of cancer (35). In clinic, up-regulation of EGFR in primary breast tumors is associated with poor prognosis (36,37). It is reported that the positive EGFR expression had a higher recurrent rate than the negative. In consistence, negative expression of EGFR had a significantly better 5-year survival rate than positive expression (38). Antibody-based immunotherapy targeting EGFR has improved the median survival of colorectal cancer patients to 24 months (39). EGFR might be a key protein in cathepsin D-promoted NPC metastasis. Our study showed that EGFR formed complexes with HSP90, which may favor the transition of HSP90 to its active conformation.
However, there was no significant association between the expression of EGFR and that of HSP90. The interplay between EGFR and HSP90 needs further explorations. EGFR and HSP90A are the key proteins during cathepsin D-mediated cell invasion and migration.
One hundred and forty-one proteins were identified by MS; we have only selectively validated two candidate biomarkers. It is necessary to look at a wider scope of each candidate protein.
Based on bioinformatics analysis and literature implications on the critical roles of cathepsin D, EGFR and HSP90A in invasion and metastasis, we further studied the interaction and correlation among cathepsin D, EGFR and HSP90A. Correlation analysis for the expression levels of cathepsin D, EGFR and HSP90A in NPC tissues indicated that the expression of cathepsin D was positively correlated with the expression of EGFR and HSP90A. Up-regulation of cathepsin D may cause up-regulation of EGFR and HSP90A, contributing to NPC metastasis. We also found that cathepsin D/EGFR/HSP90A could form complex in NPC cells. In order to further confirm that cathepsin D has an effect on EGFR and HSP90A, and that cathepsin D/EGFR/HSP90A complex has an effect on NPC invasion, we established both cathepsin D-down-regulated 5-8F NPC cell line and cathepsin D-up-regulated 6-10B NPC cell line. We examined the modulation of cathepsin D on the expression of EGFR and HSP90A, and performed in vitro invasion assay in NPC cells. The results showed that cathepsin D up-regulation increased the expression of EGFR and HSP90A, and enhanced cell invasiveness in non-metastatic 6-10B cells. On the other hand, cathepsin D down-regulation decreased the expression of EGFR and HSP90A, and weakened the cell invasiveness in NPC cells. Taken together, the results suggested that cathepsin D enhanced the in vitro invasive ability of NPC cells possibly through orchestra with EGFR and HSP90A mediated signaling pathways.
Conclusions
In this report, we employed MS-based proteomics, followed by bioinformatics analysis, and successfully identified 141 cathepsin D interaction proteins. We documented that cathepsin D regulated NPC invasion and metastasis by interacting with protein EGFR, HSP90A. Cathepsin D/EGFR/ HSP90A were closely correlated with the NPC invasion and metastasis. Up-regulation of cathepsin D led to up-regulation of EGFR and HSP90A in NPC, which enhanced the invasion ability of NPC cells. This study shed light into the molecular mechanisms of cathepsin D mediated NPC invasion and migration.
Full table
Acknowledgments
Thank Yufang Yin for reviewing the paper.
Funding: The National Science Foundation of China (contract grant number: 81372894, 81072198, 81172210, 81272959); The Scientific Natural Research Fund of Hunan Provincial Education Department (contract grant number: 10A104); The Natural Sciences Foundation of Hunan Province (contract grant number: 10JJ6035, 12JJ6080); The Construct Program of the Key Discipline in Hunan Province (contract grant number: 2011-76); China Postdoctoral Science Foundation (contract grant number: 2012M521528). The open fund of Chinese Hunan Provincial Education Department innovation platform: 15K109; The Hunan Province Science foundation: 2016JJ2012.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.10.48). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Twu CW, Wang WY, Chen CC, et al. Metronomic adjuvant chemotherapy improves treatment outcome in nasopharyngeal carcinoma patients with postradiation persistently detectable plasma Epstein-Barr virus deoxyribonucleic acid. Int J Radiat Oncol Biol Phys 2014;89:21-9. [Crossref] [PubMed]
- Laantri N, Attaleb M, Kandil M, et al. Human papillomavirus detection in moroccan patients with nasopharyngeal carcinoma. Infect Agent Cancer 2011;6:3. [Crossref] [PubMed]
- Maxwell JH, Kumar B, Feng FY, et al. HPV-positive/p16-positive/EBV-negative nasopharyngeal carcinoma in white North Americans. Head Neck 2010;32:562-7. [PubMed]
- Guo R, Chen XZ, Chen L, et al. Comorbidity predicts poor prognosis in nasopharyngeal carcinoma:Development and validation of a predictive score model. Radiother Oncol 2015;114:249-56. [Crossref] [PubMed]
- Singh N, Das P, Datta Gupta S, et al. Prognostic significance of extracellular matrix degrading enzymes-cathepsin L and matrix metalloproteases-2 [MMP-2] in human pancreatic cancer. Cancer Invest 2013;31:461-71. [Crossref] [PubMed]
- Ungefroren H, Sebens S, Seidl D, et al. Interaction of tumor cells with the microenvironment. Cell Commun Signal 2011;9:18. [Crossref] [PubMed]
- Benes P, Vetvicka V, Fusek M. Cathepsin D--many functions of one aspartic protease. Crit Rev Oncol Hematol 2008;68:12-28. [Crossref] [PubMed]
- Sharma S, Lee A, Choi K, et al. Biomimetic aggrecan reduces cartilage extracellular matrix from degradation and lowers catabolic activity in ex vivo and in vivo models. Macromol Biosci 2013;13:1228-37. [Crossref] [PubMed]
- Sun H, Lou X, Shan Q, et al. Proteolytic characteristics of cathepsin D related to the recognition and cleavage of its target proteins. PLoS One 2013;8:e65733 [Crossref] [PubMed]
- Qin AP, Zhang HL, Qin ZH. Mechanisms of lysosomal proteases participating in cerebral ischemia-induced neuronal death. Neurosci Bull 2008;24:117-23. [Crossref] [PubMed]
- Brojatsch J, Lima H Jr, Palliser D, et al. Distinct cathepsins control necrotic cell death mediated by pyroptosis inducers and lysosome-destabilizing agents. Cell Cycle 2015;14:964-72. [Crossref] [PubMed]
- Koch S, Scifo E, Rokka A, et al. Cathepsin D deficiency induces cytoskeletal changes and affects cell migration pathways in the brain. Neurobiol Dis 2013;50:107-19. [Crossref] [PubMed]
- Fonović M, Turk B. Cysteine cathepsins and their potential in clinical therapy and biomarker discovery. Proteomics Clin Appl 2014;8:416-26. [Crossref] [PubMed]
- Abdou AG, Maraee AH, Shoeib MA, et al. Cathepsin D expression in chronic plaque psoriasis: an immunohistochemical study. Acta Dermatovenerol Croat 2011;19:143-9. [PubMed]
- Knopfová L, Beneš P, Pekarčíková L, et al. c-Myb regulates matrix metalloproteinases 1/9, and cathepsin D: implications for matrix-dependent breast cancer cell invasion and metastasis. Mol Cancer 2012;11:15. [Crossref] [PubMed]
- Mohamadzadeh M, Mohamadzadeh H, Brammer M, et al. B. Identification of proteases employed by dendritic cells in the processing of protein purified derivative (PPD). J Immune Based Ther Vaccines 2004;2:8. [Crossref] [PubMed]
- Dumartin L, Whiteman HJ, Weeks ME, et al. AGR2 is a novel surface antigen that promotes the dissemination of pancreatic cancer cells through regulation of cathepsins B and D. Cancer Res 2011;71:7091-102. [Crossref] [PubMed]
- Wang JS, Wang FB, Zhang QG, et al. Enhanced expression of Rab27A gene by breast cancer cells promoting invasiveness and the metastasis potential by secretion of insulin-like growth factor-II. Mol Cancer Res 2008;6:372-82. [Crossref] [PubMed]
- Liu Z, Chen C, Yang H, et al. Proteomic features of potential tumor suppressor NESG1 in nasopharyngeal carcinoma. Proteomics 2012;12:3416-25. [Crossref] [PubMed]
- Smith B, Randle D, Mezencev R, et al. Camalexin-induced apoptosis in prostate cancer cells involves alterations of expression and activity of lysosomal protease cathepsin D. Molecules 2014;19:3988-4005. [Crossref] [PubMed]
- Cheng AL, Huang WG, Chen ZC, et al. Identification of novel nasopharyngeal carcinoma biomarkers by laser capture microdissection and proteomic analysis. Clin Cancer Res 2008;14:435-45. [Crossref] [PubMed]
- Cheng AL, Huang WG, Chen ZC, et al. Identificating cathepsin D as a biomarker for differentiation and prognosis of nasopharyngeal carcinoma by laser capture microdissection and proteomic analysis. J Proteome Res 2008;7:2415-26. [Crossref] [PubMed]
- Binder RJ. Functions of heat shock proteins in pathways of the innate and adaptive immune system. J Immunol 2014;193:5765-71. [Crossref] [PubMed]
- Turturici G, Tinnirello R, Sconzo G, et al. Positive or negative involvement of heat shock proteins in multiple sclerosis pathogenesis: an overview. J Neuropathol Exp Neurol 2014;73:1092-106. [Crossref] [PubMed]
- Treweek TM, Meehan S, Ecroyd H, et al. Small heat-shock proteins: important players in regulating cellular proteostasis. Cell Mol Life Sci 2015;72:429-51. [Crossref] [PubMed]
- Wick G, Jakic B, Buszko M, et al. The role of heat shock proteins in atherosclerosis. Nat Rev Cardiol 2014;11:516-29. [Crossref] [PubMed]
- Mjahed H, Girodon F, Fontenay M, et al. Heat shock proteins in hematopoietic malignancies. Exp Cell Res 2012;318:1946-58. [Crossref] [PubMed]
- Lee SS, Tsai CH, Ho YC, et al. Heat shock protein 27 expression in areca quid chewing-associated oral squamous cell carcinomas. Oral Dis 2012;18:713-9. [Crossref] [PubMed]
- Ischia J, Saad F, Gleave M. The promise of heat shock protein inhibitors in the treatment of castration resistant prostate cancer. Curr Opin Urol 2013;23:194-200. [Crossref] [PubMed]
- Connor AE, Baumgartner RN, Baumgartner KB, et al. Epidermal growth factor receptor (EGFR) polymorphisms and breast cancer among Hispanic and non-Hispanic white women: the Breast Cancer Health Disparities Study. Int J Mol Epidemiol Genet 2013;4:235-49. [PubMed]
- Cui J, Hu YF, Feng XM, et al. EGFR inhibitors and autophagy in cancer treatment. Tumour Biol 2014;35:11701-9. [Crossref] [PubMed]
- Kalia M. Biomarkers for personalized oncology: recent advances and future challenges. Metabolism 2015;64:S16-21. [Crossref] [PubMed]
- Hendifar A, Tan CR, Annamalai A, et al. Biomarker-driven EGFR therapy improves outcomes in patients with metastatic colorectal cancer. Expert Rev Anticancer Ther 2014;14:1051-61. [Crossref] [PubMed]
- Cheng L, Ren W, Xie L, et al. Anti-EGFR MoAb treatment in colorectal cancer: limitations, controversies, and contradictories. Cancer Chemother Pharmacol 2014;74:1-13. [Crossref] [PubMed]
- Simpson DR, Mell LK, Cohen EE. Targeting the PI3K/AKT/mTOR pathway in squamous cell carcinoma of the head and neck. Oral Oncol 2015;51:291-8. [Crossref] [PubMed]
- Guo B, Gao J, Zhan J, et al. Kindlin-2 interacts with and stabilizes EGFR and is required for EGF-induced breast cancer cell migration. Cancer Lett 2015;361:271-81. [Crossref] [PubMed]
- Bliesath J, Huser N, Omori M, et al. Combined inhibition of EGFR and CK2 augments the attenuation of PI3K-Akt-mTOR signaling and the killing of cancer cells. Cancer Lett 2012;322:113-8. [Crossref] [PubMed]
- Cao XJ, Hao JF, Yang XH, et al. Prognostic value of expression of EGFR and nm23 for locoregionally advanced nasopharyngeal carcinoma. Med Oncol 2012;29:263-71. [Crossref] [PubMed]
- Merla A, Goel S. Novel drugs targeting the epidermal growth factor receptor and its downstream pathways in the treatment of colorectal cancer: a systematic review. Chemother Res Pract 2012;2012:387172 [Crossref] [PubMed]


