
Novel mechanisms of chemoresistance by Fusobacterium nucleatum involve not so novel pathways of microRNAs and autophagy
Colon cancer is the second leading cause of cancer death among both men and women in the United States. Even with available chemotherapies, a significant number of patients have recurrent disease, leading to the high incidence of morbidity and mortality. Understanding the mechanisms by which chemoresistance can occur is paramount to developing novel therapeutic approaches. Multiple past studies have highlighted the modes of resistance in cancer cells. The primary focus has been on molecular pathways and signaling cascades involving growth factors, tyrosine kinases and transcription factors that lead to tumorigenesis as well as chemoresistance (1,2). In the past few years, microRNAs (miRNAs) and small interfering-RNA (siRNA) have gained a significant role in cancer biology.
In addition to the intrinsic factors, we cannot ignore the environmental factors that impact these cellular mechanisms. In fact, there is a remarkable burden of infection-related malignancies worldwide. It is now well-established that oncoviruses such as human papilloma virus (HPV) and Epstein-Barr virus (EBV) can cause primary malignancies. Similarly, bacteria such as Helicobacter pylori have been implicated in gastric cancers; and parasites like Clonorchis and Opisthorchis are known to cause cholangiocarcinoma. These organisms lead to cancer either by altering host genome (as in oncoviruses), or by deregulation of signaling pathways via inflammatory processes of the host cells (3).
In the past few years, there is a developing role for Fusobacterium nucleatum (F. nucleatum) in cancer treatment. F. nucleatum is an anaerobic, gram-negative bacterium that is primarily found in the oral cavity of primates. Even though they are commensal organisms, they have been implicated in periodontal diseases as well as in obstetric complications (4). The bacterium has been studied in inflammatory bowel disease (IBD), and more recently in the pathogenesis of colorectal cancer (CRC). Initial observational studies had showed that Fusobacterium, along with Coriobacteriaceae, Roseburia, and Faecalibacterium were overrepresented in colon cancer cells and regarded as “passenger” organisms, without any implication in carcinogenesis (5). Additional genomic studies of the microbiome associated with CRC revealed that there was significant enrichment of Fusobacterium in the cancer itself and not the surrounding normal tissues (6). Chronic exposure of APC (Min/+) mice with human F. nucleatum isolate showed increased colon tumor burden. Additional studies by multiple groups (7-9) reported that that this organism may in fact potentiate intestinal tumorigenesis and act as a modulator in tumor-immune microenvironment. The proposed molecular mechanisms included interaction between Fusobacterium FadA adhesin and host cadherins (8); and Fusobacterium lectin Fap2 and host Gal-GalNAc (9), which stimulates oncogenic beta-catenin signaling pathway that can lead to cancer. A recent study reported a direct correlation between F. nucleatum and proliferation, invasive activity, and xenograft tumor formation in mice, in addition to implicating miRNA-21 as a key factor in the molecular pathway (10).
MicroRNAs (miRNAs) are evolutionarily conserved, short non-coding RNAs 20–24 nucleotides in length that play important roles in virtually all biological pathways. There are approximately 1,400 miRNAs found in humans, and it is estimated that at least 60% of human protein-coding genes are regulated by miRNAs. They are initially transcribed as primary transcripts (pri-mRNAs) by RNA polymerase II, from either introns or exons of protein coding genes, in addition to intergenic regions (11). Once transcribed, the microprocessor complex containing the RNAse III enzyme Drosha cleaves the primary transcript to release the pre-miRNA hairpin that is subsequently exported to the cytoplasm via exportin 5 (XPO5). In the cytoplasm, a protein complex that includes DICER, another RNAase III enzyme, and transactivation-responsive RNA-binding protein (TRBP) further process pre-miRNA to produce double-stranded mature miRNA, which subsequently incorporates into the RNA-induced silencing complex (RISC) where the passenger (miRNA*) strand is selectively degraded. Along with Argonaute 2 (AGO2) and other RISC factors, the mature miRNAs bind to complementary sites on messenger RNA transcripts to induce either translational pausing or transcript degradation.
In an article recently published in the journal Cell (12), the authors highlight that autophagy and its regulation by miRNA is an important step in cancer chemoresistance. Autophagy is a highly regulated, cellular catabolic process by which dysfunctional cellular components are degraded to maintain homeostasis. The process was initially thought to involve only cell death, but more recent studies have shown that a major function is to maintain cell survival under stressful conditions such as nutrient deprivation, hypoxia, reactive oxygen species, chemotherapy, DNA damage or even intracellular pathogens that would otherwise lead to cell death. Physiologically, this process plays a vital role in turnover of cellular components, clearance of intracellular microbes, and even in the regulation of innate and adaptive immunity. The mechanistic and molecular pathway for autophagy is complex, but well conserved across eukaryotic organisms. AuTophaGy-related (ATG) proteins are encoded by 31 highly conserved genes, and have specific functions along the autophagy mechanistic pathway (13). Homologous proteins to ATGs are present in mammals, such as UNC-51-like Kinases (ULK) proteins.
The autophagy process has mechanistically distinct steps, including (I) autophagy induction and phagophore formation; (II) vesicle nucleation; (III) vesicle elongation and completion; (IV) retrieval; and (V) fusion between autophagosomes and lysosomes (Figure 1) (14). During the induction phase, autophagy is initiated by protein complexes composed of ULK1/2-ATG13-FIP200-ATG101; and inhibited by mTORC1 (mammalian target of rapamycin) complex 1. MiRNA miR-885-3P has been shown to play a role in regulation of this phase (15). Another study showed a direct role of ULK1 in AML differentiation and its regulation by miRNA-106a (16). The miRNAs and mTOR pathways, which have essential function in autophagy, have also been shown to have bidirectional mechanisms, which suggests that the degree of regulation in various molecular pathways is much more significant than previously thought (17). The study by Yu et al. (12) shows that ULK1 gene is downregulated by miRNA miR-18a*.
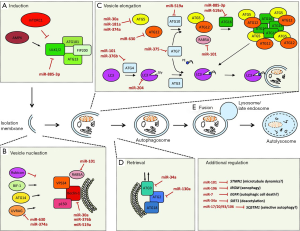
The vesicle nucleation phase is the first step in which proteins and lipids are recruited for formation of the autophagosomal membrane. This involves the activation Beclin-1/Class III PI3K complex and subsequent recruitment of additional ATGs. The pathways of autophagy and miRNAs have paralleled each other in the past two decades. In 2009, the two were linked when BECN1, which codes for the Beclin-1 autophagy-promoting gene in some cancer cells, was shown to be modulated by miR-30a (18). Later studies then reported that Beclin-1 was regulated by multiple other miRNAs, including miR-376b (19) and miR-519a (20).
In the next step of autophagy, the phagophore elongates into a double membrane organelle, requiring two pathways for ubiquitin-like conjugation. One pathway involves the covalent conjugation of ATG12-ATG5, which requires Ubiquitin-activating E1- and E2-like enzymes ATG7 and ATG10, respectively. The second pathway involves the conjugation of microtubule-associated protein light chain 3 (LC3B) to phosphatidylethanolamine (PE) and generation of LC3B-II, which requires ATG7, E2-like enzyme ATG3, and another ATG protein complex. LC3II is anchored to both inner and outer faces of the phagophore membrane and is a commonly used experimental marker of autophagy. ATG7 has also been identified as a direct target of miRNA-375 in hepatocellular carcinoma (21). In the Cell study (12), ATG7 was shown to be regulated by miR-4802, whose function had not been previously described.
The retrieval stage in autophagy involves the recruitment of additional proteins and lipids, mostly involving ATG9. The molecular mechanism(s) as it relates to other pathway stages is unclear (21) and the majority of our understanding of this step is derived from yeast studies. The final step in the autophagic process is the fusion of the autophagosome into the lysosomes. This involves proteins such as LAMP2 and RAB7, resulting in vesicle breakdown and cargo degradation in autolysosomes by lysosomal hydrolases. No miRNAs have been identified to have a direct role in fusion, but computational studies suggest otherwise.
Yu et al. (12) highlight a series of experiments that demonstrate a relationship between F. nucleatum, recurrent CRC, and the role of the organism in mediating the chemoresistance to chemotherapeutic agents against CRC. Furthermore, the molecular mechanism(s) involving miRNA and autophagy is elaborated via well-designed experimental studies. Until this study, the role of F. nucleatum in chemoresistance and subsequent recurrence of cancer had not been previously reported.
The authors initially confirmed the relationship between F. nucleatum and recurrent CRC by showing that the number of organisms in tissues was higher compared to non-recurrent samples. The authors then explore the clinicopathological significance of this finding, which predicted reduced recurrent-free-survival of the patient, cancer aggressiveness, in addition to the prediction of recurrence of the cancer. These observations were then validated in a third cohort of patients with colon cancer.
The authors then designed a series of experiments to test the hypothesis in which increased concentration of F. nucleatum was the cause rather than effect of cancer recurrence as well as chemoresistance. The gene expression profiles in CRC cells that were cultured with F. nucleatum showed significantly increased expression of ATG7 and ULK1 mRNA, which led the authors to postulate that the autophagy pathway for cell survival may be activated by F. nucleatum. Functional assays were performed, and showed increased LC3-II (autophagosomal marker LC3-II reflects starvation-induced autophagic activity, and its detection by immunoblotting or immunofluorescence has become a reliable method for monitoring autophagy and autophagy-related processes) and decreased p62 expressions (p62 accumulates when autophagy is inhibited, and decreased levels can be observed when autophagy is induced), thus establishing that F. nucleatum is implicated for inducing autophagy.
To establish the relationship between autophagy and chemoresistance, the authors co-cultured CRC cells with F. nucleatum and treated these cells with Oxaliplatin and 5-Fluorouracil (common drugs used in treatment of CRC). They noted inhibition of cleavage of proteins such as caspases, PARP and p-H2AX in these cells, therefore avoiding apoptosis and leading to cell survival. In these cells, ULK1 and ATG7 were shown to be upregulated. When the cancer cells were transfected with ULK1-siRNA or ATG7-siRNA, the inhibition of apoptosis was abolished, essentially negating the effect of F. nucleatum in the cancer cells, and showing that these two elements are upregulated by F. nucleatum. However, recombinant reporter plasmids containing the promoter regions of the autophagy elements ATG7 or ULK1 showed no transcriptional activity, suggesting that increases in mRNA levels were not the result of transcriptional activation.
Further, prompted by work from recent studies showing a role for miRNA in biologic pathways, miRNA expression profiling on these tissues was done which showed that there was selective downregulation of 68 miRNAs. Among those, miR-18a* and miR-4802 were found to have key roles in regulation of the autophagy elements ATG7 and ULK1. These miRNAs would have otherwise targeted the seed regions within 3’-UTR of the ULK1 and ATG genes to decrease their RNA levels. When the mimics of these miRNAs were transfected into the CRC cell cultures with F. nucleatum, they increased chemotherapy-induced apoptosis in the cells. These mimic miRNAs did so by abolishing the inhibitory effects that the organism had on the chemotherapy-induced cleavage of the caspases, PARP and p-H2AX proteins. Further experiments on xenograft mouse models confirmed these findings.
The next step of experiments was to examine the host-microbe interaction. TLR4 and MYD88 innate immune signaling pathways were activated in cells treated with F. nucleatum, and these findings were also seen in prior studies. The proposed mechanism was that F. nucleatum activates the TLR4 and MYD88 pathway which selectively downregulates miRNA miR-18a* and miRNA-4802, and in doing so, activates the autophagy pathway, steering the cell away from apoptosis, and consequently increasing chemoresistance in cancer cells.
The role of miRNA is gaining significant interest in the field of oncogenesis. Alterations in miRNA genes by various mechanisms, including overexpression, deletions or mutations have been demonstrated to have causal relationship in cancer development. The downhill effects of these alterations impact various protein-coding oncogenes or tumor suppressor genes, which then eventually leads to cancer initiation and/or proliferation (22). In chronic lymphocytic leukemia, the role of miRNA in cancer proliferation has been well studied (23). A large-scale miRnome analysis on multiple solid tumors identified miRNA signature profiles composed by a large number of overexpressed miRNAs (24). Numerous studies have been published in the past several years that have explored the roles of miRNA in various pathways of colon cancer proliferation and suppression (25). In Yu et al. (12), the authors note that miR-18a* acts on cellular mechanisms for the development of chemoresistance via modulation of autophagy. Interestingly, this specific miRNA miR-18a* has been characterized in prior studies as a potential tumor suppressor by targeting K-Ras (26), as well as inhibitor of tumor suppressor CDC42 in CRC cells (27). In contrast, in one study miR-18a was shown to promote malignant progression by impairing miRNA biogenesis in nasopharyngeal carcinoma (28). These findings suggested that miRNAs have diverse functions and potentially regulate multiple pathways (11).
Another layer of complexity to chemoresistance and the role of autophagy is added when one considers the microbiome. Human gut harbors approximately 10–100 trillion microorganisms, which includes 100–200 different bacterial species, and 2–4 million genes. A recent study highlighted the role of miRNA in gut microbiome-host communication, which showed that fecal miRNA plays an important role in bacterial gene expressions, and affects gut microbial growth and composition (29). Earlier this year, a study identified the role of F. nucleatum-induced miRNAs and its role in colon cancer (10). The authors reported that TLR4 and MYD88 were involved in activation of NF-κB and increased expression of miR21; and this miRNA reduced the levels of RAS GTPase RASA1, leading to colon cancer proliferation. Yu et al. (12) elegantly showed that F. nucleatum not only plays a role in proliferation and recurrence of cancer, but also plays a key role in the development of chemoresistance via miRNA-mediated regulation of autophagy. The role of molecular pathways for chemoresistance that involve miRNAs and autophagy is promising, and certainly exciting for the discovery of novel targets in the treatment of CRC. However, translation from bench-to-bedside poses many challenges.
The relationship between CRC and Fusobacterium was initially observational, and it is not entirely clear as to what factors are involved in their recruitment to influence the progression of colon cancer. It is well known that cancer cells can evade the immune system for growth. It would be an interesting study to see if there are “unsung heroes” in the gut microbiota that keep colon cancer in check, and even more interesting, if there are mechanisms that cancer cells use to evade the “unsung hero” organisms. Host shaping of the gut flora with miRNA (29) is an intriguing approach to therapy. Future studies could also focus on profiling fecal miRNA in colon cancer cells and their role in modifying the microbiota profile for cancer development and evasion from chemotherapy. Without addressing some of these questions, it is unclear from the current study that targeting one organism as a therapeutic method will be beneficial in cancer recurrence prevention. This elegant and rigorously done study is promising for the use of F. nucleatum as a screening tool for colon cancer recurrence. However, the negative predictive value and sensitivity of bacterial monitoring would have to be taken into careful consideration before it can be used as an effective tool, and this would require wider population based studies.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Fei Pan (Department of Gastroenterology and Hepatology, Division of Internal Medicine, PLA Medical School & PLA General Hospital, Beijing, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.12.20). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Abdullah LN, Chow EK. Mechanisms of chemoresistance in cancer stem cells. Clin Transl Med 2013;2:3. [Crossref] [PubMed]
- Pommier Y, Sordet O, Antony S, et al. Apoptosis defects and chemotherapy resistance: molecular interaction maps and networks. Oncogene 2004;23:2934-49. [Crossref] [PubMed]
- van Tong H, Brindley PJ, Meyer CG, et al. Parasite infection, carcinogenesis and human malignancy. EBioMedicine 2017;15:12-23. [Crossref] [PubMed]
- Han YW.
Fusobacterium nucleatum: a commensal-turned pathogen. Curr Opin Microbiol 2015;23:141-7. [Crossref] [PubMed] - Marchesi JR, Dutilh BE, Hall N, et al. Towards the human colorectal cancer microbiome. PLoS One 2011;6:e20447 [Crossref] [PubMed]
- Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of
Fusobacterium with colorectal carcinoma. Genome Res 2012;22:292-8. [Crossref] [PubMed] - Kostic AD, Chun E, Robertson L, et al.
Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013;14:207-15. [Crossref] [PubMed] - Rubinstein MR, Wang X, Liu W, et al.
Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 2013;14:195-206. [Crossref] [PubMed] - Abed J, Emgård JE, Zamir G, et al. Fap2 mediates
Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe 2016;20:215-25. [Crossref] [PubMed] - Yang Y, Weng W, Peng J, et al.
Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating Toll-like receptor 4 signaling to nuclear factor-κB, and up-regulating expression of miRNA-21. Gastroenterology 2017;152:851-66. [Crossref] [PubMed] - Subramanian S, Steer CJ. MicroRNAs as gatekeepers of apoptosis. J Cell Physiol 2010;223:289-98. [PubMed]
- Yu T, Guo F, Yu Y, et al.
Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 2017;170:548-63.e16. [Crossref] [PubMed] - He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 2009;43:67-93. [Crossref] [PubMed]
- Frankel LB, Lund AH. MicroRNA regulation of autophagy. Carcinogenesis 2012;33:2018-25. [Crossref] [PubMed]
- Huang Y, Chuang AY, Ratovitski EA. Phospho-ΔNp63α/miR-885-3p axis in tumor cell life and cell death upon cisplatin exposure. Cell Cycle 2011;10:3938-47. [Crossref] [PubMed]
- Tschan MP, Jost M, Batliner J, et al. The autophagy gene ULK1 plays a role in AML differentiation and is negatively regulated by the oncogenic miRNA-106a. Blood 2010;116:223.
- Zhang Y, Huang B, Wang HY, et al. Emerging role of microRNAs in mTOR signaling. Cell Mol Life Sci 2017;74:2613-25. [Crossref] [PubMed]
- Zhu H, Wu H, Liu X, et al. Regulation of autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer cells. Autophagy 2009;5:816-23. [Crossref] [PubMed]
- Korkmaz G, le Sage C, Tekirdag KA, et al. miR-376b controls starvation and mTOR inhibition-related autophagy by targeting ATG4C and BECN1. Autophagy 2012;8:165-76. [Crossref] [PubMed]
- Huang Y, Guerrero-Preston R, Ratovitski EA. Phospho-ΔNp63α-dependent regulation of autophagic signaling through transcription and micro-RNA modulation. Cell Cycle 2012;11:1247-59. [Crossref] [PubMed]
- Chang Y, Yan W, He X, et al. miR-375 inhibits autophagy and reduces viability of hepatocellular carcinoma cells under hypoxic conditions. Gastroenterology 2012;143:177-87.e8. [Crossref] [PubMed]
- Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol 2012;6:590-610. [Crossref] [PubMed]
- Balatti V, Pekarky Y, Croce CM. Role of microRNA in chronic lymphocytic leukemia onset and progression. J Hematol Oncol 2015;8:12. [Crossref] [PubMed]
- Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 2006;103:2257-61. [Crossref] [PubMed]
- Schetter AJ, Okayama H, Harris CC. The role of microRNAs in colorectal cancer. Cancer J 2012;18:244-52. [Crossref] [PubMed]
- Tsang WP, Kwok TT. The miR-18a* microRNA functions as a potential tumor suppressor by targeting on K-Ras. Carcinogenesis 2009;30:953-9. [Crossref] [PubMed]
- Humphreys KJ, McKinnon RA, Michael MZ. miR-18a inhibits CDC42 and plays a tumour suppressor role in colorectal cancer cells. PLoS One 2014;9:e112288 [Crossref] [PubMed]
- Luo Z, Dai Y, Zhang L, et al. miR-18a promotes malignant progression by impairing microRNA biogenesis in nasopharyngeal carcinoma. Carcinogenesis 2013;34:415-25. [Crossref] [PubMed]
- Liu S, da Cunha AP, Rezende RM, et al. The host shapes the gut microbiota via fecal microRNA. Cell Host Microbe 2016;19:32-43. [Crossref] [PubMed]


