
Identification of potential gene and microRNA biomarkers for colon cancer by an integrated bioinformatical approach
Introduction
The NCI-60 is a panel of 60 human tumor cell lines which isolated from diverse histologies and 9 different tissues of origin. It mainly contains cancers of colorectal (CO), ovarian (OV), melanomas (ME), lung (LC), breast (BR), and central nervous system (CNS) origin (1). The NCI-60 panel was used by the Developmental Therapeutics Program (DTP) of the National Cancer Institute (NCI) of the USA in 1990. It is reported that NCI-60 is a comprehensively panel of diverse cell types at the DNA, RNA, protein, mutation, functional, and pharmacological levels (2-5). Now NCI-60 has been widely used in cancer research and bioinformatics for analysis cell phenotypes and pathway relationships (6).
Microarray is a powerful tool for detecting the expression pattern of RNA levels, including those of mRNA and microRNAs (miRNA) (7,8). Due to the wide use of microarray, a large number of microarray data have been collected and used to discover mechanism for tumorigenesis, development and therapy. Bioinformatical approaches are crucial for discovering more valuable information included in these datasets, particularly signaling pathways, complex biological processes and the interaction network of differentially expressed genes (DEGs) and differentially expressed miRNAs (DEMs) (9,10).
Colon cancer is the fourth-leading cause of cancer-related death in the world. Despite after surgical resection of colon cancer, more than 50% of patients die of developing recurrence and disease relapse after several months (11,12). Clinical diagnosis of colon cancer relatively lagged behind dues to a serious threat to human health. Therefore, it is necessary to discover more effective biomarkers and reveal prediction molecular mechanisms of clinical significance of biomarkers.
Recently, construction of biophysics network models of numerous components has contributed to our understanding of the relationship between different types of biological molecules (13). In this study, we used the simple Pearson correlation coefficient (PCC) of the gene and miRNA expression profile over diverse NCI-60 cell lines. It is possible to obtain a certain number of significant correlations between gene expression and miRNAs on subsets of cancer samples.
In this research, our goal is to identify the DEGs and DEMs for colon cancer, functional enrichment analysis for DEGs, construct miRNA network with their target genes and gene-miRNA correlation network, which provide new clues on possible mechanisms for colon cancer.
Methods
Acquisition of microarray data
Microarray gene expression and miRNA expression data were obtained from the NCBI Gene Expression Omnibus (GEO) ftp (https://www.ncbi.nlm.nih.gov/geo/). NCI-60 gene expression data (GSE32474) and miRNA expression data (GSE26375) were retrieved. Gene probes or miRNAs with a fold change of >4 and P<0.01 for colon cancer over the median value were selected for the further analysis.
Functional analysis and pathway enrichment analysis
DAVID (http://david.ncifcrf.gov/) is an online tool that can be utilized to perform a functional analysis and pathway enrichment analysis for discovering the relationships among the selected gene sets (14). Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was performed by the DAVID online program.
Analysis of miRNA-target mRNA network
The selected DEMs were imported to TargetScan (Release 7.1, http://www.targetscan.org/) which can search the target genes based on the conserved sites matching the seed regions of miRNAs. Cumulative weighted context++ score ≤−0.4 were set as the threshold. The miRNA-target mRNA network was constructed by Cytoscape (http://www.cytoscape.org/) (15).
Constructing gene-miRNA network
PCC and its P value between genes and miRNAs were calculated using gene and miRNA expression data from NCI-60 cell lines. The criteria of PCC >0.7 or PCC <−0.7 and P value <0.01 were applied to select gene-micorRNA relationship for further analysis. Finally, the network was constructed for links between compounds and genes using Cytoscape (http://www.cytoscape.org/) (15). The force-layout algorithm was applied to optimize the topology of the network.
Cell culture
HCT-116, HCT-8, A549 and H460 cell lines were all purchased from Cell Bank of Shanghai Institute of Biochemistry. HCT-116 and HCT-8 cells were cultured in DMEM/High Glucose (HyClone) medium containing 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 mg/L streptomycin. A549 and H460 cells were cultured in RPMI 1640 medium (HyClone) with 10% FBS, 100 U/mL penicillin, and 100 mg/L streptomycin. All the cells were cultured under 37 °C and 5% CO2.
Quantitative real-time PCR
Total RNA was extracted from cells and tissues using TRI Reagent RT (Invitrogen, Carlsbad, CA, USA). Total RNA solution was stored at −70 °C. One µg total RNA was converted to cDNA using Stem-loop RT primers for miRNA and using anchored oligo (dT) primers for the gene. Following the manufacturer’s instructions, 2 µL cDNA was performed to real-time qPCR using SYBR® Green PCR Kit. Beta-actin was used as a loading control for gene while U6 for miRNA. The sequences of primers used in qRT-PCR were as showed in Table 1. All reactions had three replicates. The data were analyzed using the comparative 2−ΔΔCT method.
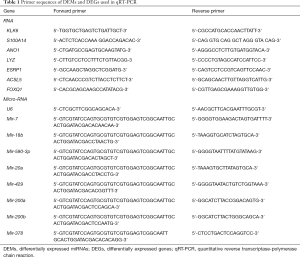
Full table
Software support and statistical analysis
Hierarchical clustering of gene expression profile was carried out using QCanvas (16). All images were formatted for optimal presentation using Adobe Illustrator CS4 (Adobe Systems, San Jose, CA, USA). To determine statistical significance, P value from t-statistic was calculated.
Results
Identification of DEGs
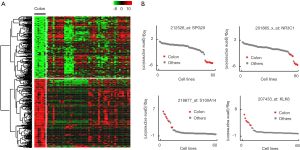
To identify novel gene biomarkers for colon cancer, the DNA microarray of the NCI-60 cell line panel (GSE32474) was analyzed. A total of 560 gene probes were identified with significant differential expression in colon cancers. Among them, 285 gene probes with higher expression level and 265 gene probes with lower expression levels in colon cancer cells [log2 (fold change) >2 or log2 (fold change) <−2, P<0.01]. The up-regulated and down-regulated genes (DEGs) were used to generate the heatmap profile (Figure 1A). The differential expression pattern for two down-regulated genes (SPG20 and NR3C1) and two up-regulated genes (S100A14 and KLK6) were showed in Figure 1B.
GO function and KEGG pathway analyses
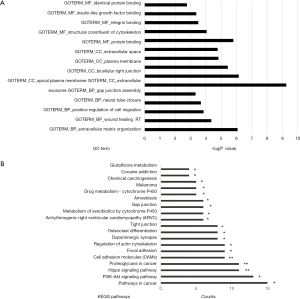
To explore the functions of those DEGs in depth, the biological process, pathway annotation and molecular function were revealed using DAVID Gene Functional Classification Tool. The GO analysis revealed that the DEGs were significantly involved in the gap junction assembly, neural tube closure, positive regulation of cell migration, wound healing and extracellular matrix organization (the top 5 biological process) (Figure 2A). Furthermore, the KEGG pathway was used to determine the pathways involved in colon cancers. The result showed that DEGs were primarily involved in pathways in cancer, PI3K-AKT signaling pathways, cell adhesion, focal adhesion, tight junction and etc. (Figure 2B). The results collectively indicated that genes involved in the cell proliferation and migration were significantly associated with colon cancers.
Experimental validation of the key DEMs
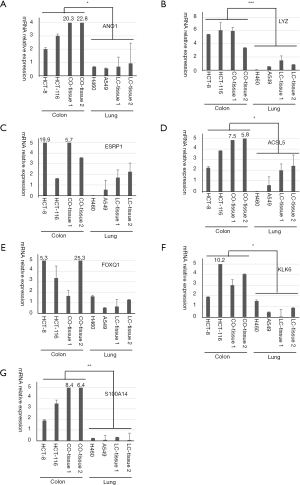
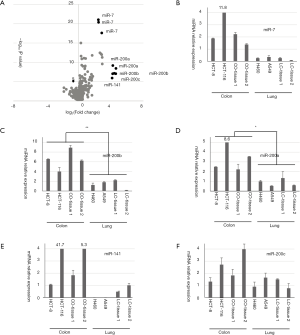
According to the transcriptomic data and bioinformatics analysis, seven genes were selected as potential gene biomarker candidates, including KLK6, S100A14, ANO1, LYZ, ESRP1, ACSLC5 and FOXQ1, and their roles in colon cancer have not been clearly addressed before. The previous study has been showed that KLK6 may represent a potential unfavorable prognostic biomarker for colon cancer (17). Overexpression of S100A14 in colon tumors may have a potentially important function in malignant transformation (18). These seven genes were further verified by qRT-PCR. As showed in Figure 3, the expression level of ANO1, LYZ, ESRP1, ACSLC5, FOXQ1, KLK6 and S100A14 were significant highly expressed in colon cancer cells and colon tissues.
Identification of DEMs and miRNA-target mRNA network
There were 27 miRNA probes were identified from GSE26375 for colon cancers [log2 (fold change) >2 or log2 (fold change) <−2, P<0.01]
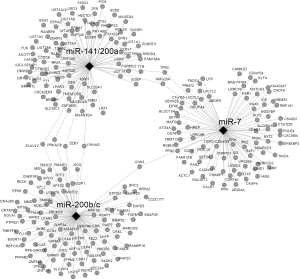
Construction of gene-miRNA network
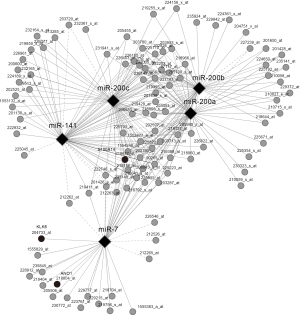
To further explore the interactions of DEGs and DEMs, the PCC and its P value between genes and miRNAs were calculated using gene expression and miRNA expression data on 59 NCI-60 cell lines. The criteria of absolute PCC >0.7 and P value <0.01 were applied to select gene-miRNA relationship for further analysis. Finally, the network for a total of 104 gene probes and 5 miRNAs was constructed using Cytoscape (Figure 6). The data provide a useful resource for studying and predicting the relationship of genes and miRNAs for colon cancer.
Discussion
Despite advance in medical and surgical therapy, the colon cancer is the third common diseases in the world, moreover, in developed countries, colon cancer has a higher incidence than developing countries (20). In developed countries, more than 65% of diseases are colon cancers (21). The characteristics of colon cancer are invasive and spread to other tissues of the human body. The lethality of lethal malignancy globally is due to the difficulties in detecting colon cancer at an early state. Therefore, it is essential and beneficial to figure out the etiological factors and mechanisms of colon cancer to improve prevention and survival rate (22). Recently, microarray technology has been an effective tool for revealing the expression of general genetic alteration in different physiological and pathological status, which enables the discovery of new targets for diagnosis, therapeutic, and prognosis of malignant cancers.
In this study, a total of 560 gene probes, including 285 significantly up-regulated genes and 265 significantly down-regulated genes were identified used NCI-60 microarray dataset. These differential expressed genes mainly were enriched in cell proliferation, migration, invasion, apoptosis and survival rate. Among those genes, seven genes were selected as potential gene biomarker candidates, including KLK6, S100A14, ANO1, LYZ, ESRP1, ACSLC5 and FOXQ1. Anoctamin-1 (ANO1) as transmembrane protein 16A (TMEM16A) was found in a number of cancers, such as gastrointestinal stromal tumor (GIST), BR cancer (23,24). Moreover, ANO1 plays an important role in cell proliferation, tumorigenesis and progression (25). Recent studies shown that in various cell types of ANO1 could activate calcium-activated chloride channels (CaCCs), molecular and electrophysiological studies indicated that ANO1 plays a critical role in metastatic tumor (26,27). Kallikrein-related peptidase (KLK) is a member of the human kallikrein gene family, in length, KLK6 encodes of 224 amino acid with trypsin-like activity (28). In addition, the KLK6 gene was validated to be a secreted protein which shares significant homologies with other kallikreins and the enzyme (29). As a new potential biomarker, the expression of KLK6 was significantly increased in patients with cancer in an late-stage, accumulating evidence demonstrates that KLK6 may play a role in the development and progression of cancer (30). S100A14 is one protein of S1004 proteins. It has been reported that play a role in cancer cells invasion and metastasis (31). S100A14 protein exert its function which regulates tumor invasion by modulating the level of matrix metalloproteinase MMP-1 and MMP-9 (32). In addition, S100A14 could regulate the expression of MMP-2 and p-53 dependent pathway to affect tumor cells invasion (33). As a member of the long-chain acyl-CoA synthetase (ACSL) gene family, ACSL5 is overexpression on tumor versus other members of ACSL family (34,35). As a unique position among the ACSL gene family, ACSL5 at chromosome 10q25.1–q25.2, mainly located on mitochondria and regulated apoptosis of cells (36). While, human ACSL5 exert activation usually through certain apoptosis pathway (37).As a factor associated with metastatic, epithelial splicing regulatory protein 1 (ESRP1), is an RNA-binding proteins (RBPs) and splicing factor (38). ESRP1 have ability to bind the 5’UTR of a wide range of cancer-related genes and alter their translation to exhibit it functions, as a tumor expression ESRP1 inhibit epithelial to mesenchymal transition (EMT) in various cancers (39-42). However, in the present study, in cancer cells over-expressing of ESRP1 cloud enhance their metastatic potential (43). Moreover, metastatic progression of carcinoma cells is a positive association with the level of CD44v6 isoform, a target of ESRP1 (44). And ESRP1 could target growth factor receptor including FGFR1/2 pathway, AKT signaling and Snail activation to promote cancer progression (38).
miRNA, a type of small non-coding RNA molecule which consists of 18–25 nucleotides. It can regulate target gene expression to affect cell biological process such as cell apoptosis, cell proliferation, cell differentiation, and cell metastatic (45). Increasing evidence demonstrated that some miRNA can affect the pathogenesis of various cancers, including colon cancer. Therefore, miRNA may be biomarker for tumor diagnosis and treatment (46). In the present study, we identified 27 DEMs for colon cancer cells. Especially, five up-regulated miRNAs (miR-7, miR-200a, miR-200b, miR-200c, and miR-141) were identified. MiRNA-7 plays a pivotal roles on tumors growth and metastatic by affecting the region of focal adhesion kinase (FAK) mRNA, which inhibit the level of FAK protein (47). In cancer cells, miRNA-7 inhibits the expression of FAK, FAK is mainly located at the cell cytoplasm and promotes the secretion of MMPs, and therefore, miRNA-7 may be targeting the level of MMPs to exert its function (48). MiRNA-200 family consisting of five members (miRNA-200a, miRNA-200b, miRNA-200c, miRNA-141 and miRNA-429) which participating in the progression of EMT (49). As a member of miRNA-200, miRNA-429 expression in colon cancer was significantly up-regulated compared with LC, as reported, miRNA-429 have the ability to inhibit loss of E-cadherin undergoing EMT (50). Similar results that, miRNA-200a and miRNA-200b were up-regulated in colon cancer cells and tissue, moreover, miRNA-200a and miRNA-200b showed higher expression in cancer tissue than normal tissue which were identified as candidate biomarkers in tumor (51,52).
In summary, data integration and data mining play critical roles for finding the candidate biomarkers and the mechanisms of cancers. In this study, we analyzed a total of 560 DEGs and 27 DEMs. Some significant miRNAs and genes were validated by qRT-PCR, including ANO1, KLK6, A100A14, ACSL5, ESRP1 genes and miRNA-7, miRNA-200b, miRNA-200a, miRNA-200c and miR-141. Nevertheless, these identified genes and miRNA need to be confirmed by more researchers in the future, our study could provide new methods for diagnosis and treatment of colon cancer patients.
Acknowledgments
Funding: This study was funded by the National Nature Science Foundation of China [81602621] and Postdoctoral Applied Research Programs of Qingdao [2016061].
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.12.09). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The clinical samples were obtained with informed consent, and the study protocol was approved by the Ethics Committee of the Affiliated Hospital of Medical College Qingdao University.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Shoemaker RH, Monks A, Alley MC, et al. Development of human tumor cell line panels for use in disease-oriented drug screening. Prog Clin Biol Res 1988;276:265-86. [PubMed]
- Nishizuka S, Charboneau L, Young L, et al. Proteomic profiling of the NCI-60 cancer cell lines using new high-density reverse-phase lysate microarrays. Proc Natl Acad Sci U S A 2003;100:14229-34. [Crossref] [PubMed]
- Weinstein JN, Pommier Y. Transcriptomic analysis of the NCI-60 cancer cell lines. C R Biol 2003;326:909-20. [Crossref] [PubMed]
- Blower PE, Verducci JS, Lin S, et al. MicroRNA expression profiles for the NCI-60 cancer cell panel. Mol Cancer Ther 2007;6:1483-91. [Crossref] [PubMed]
- Reinhold WC, Varma S, Sousa F, et al. NCI-60 whole exome sequencing and pharmacological CellMiner analyses. PLoS One 2014;9:e101670 [Crossref] [PubMed]
- Reinhold WC, Sunshine M, Varma S, et al. Using CellMiner 1.6 for Systems Pharmacology and Genomic Analysis of the NCI-60. Clin Cancer Res 2015;21:3841-52. [Crossref] [PubMed]
- Shirota Y, Kaneko S, Honda M, et al. Identification of differentially expressed genes in hepatocellular carcinoma with cDNA microarrays. Hepatology 2001;33:832-40. [Crossref] [PubMed]
- Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005;433:769-73. [Crossref] [PubMed]
- Storey JD, Tibshirani R. Statistical methods for identifying differentially expressed genes in DNA microarrays. Methods Mol Biol 2003;224:149-57. [PubMed]
- Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 2006;103:2257-61. [Crossref] [PubMed]
- Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin 2016;66:271-89. [Crossref] [PubMed]
- Siegel R, DeSantis C, Virgo K, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin 2012;62:220-41. [Crossref] [PubMed]
- Pe'er D, Hacohen N. Principles and strategies for developing network models in cancer. Cell 2011;144:864-73. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Kim N, Park H, He N, et al. QCanvas: An Advanced Tool for Data Clustering and Visualization of Genomics Data. Genomics Inform 2012;10:263-5. [Crossref] [PubMed]
- Vakrakou A, Devetzi M, Papachristopoulou G, et al. Kallikrein-related peptidase 6 (KLK6) expression in the progression of colon adenoma to carcinoma. Biol Chem 2014;395:1105-17. [Crossref] [PubMed]
- Pietas A, Schluns K, Marenholz I, et al. Molecular cloning and characterization of the human S100A14 gene encoding a novel member of the S100 family. Genomics 2002;79:513-22. [Crossref] [PubMed]
- Humphries B, Yang C. The microRNA-200 family: small molecules with novel roles in cancer development, progression and therapy. Oncotarget 2015;6:6472-98. [Crossref] [PubMed]
- Moriarity A, O'Sullivan J, Kennedy J, et al. Current targeted therapies in the treatment of advanced colorectal cancer: a review. Ther Adv Med Oncol 2016;8:276-93. [Crossref] [PubMed]
- McGuire S. World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press, 2015. Adv Nutr 2016;7:418-9. [Crossref] [PubMed]
- Sharif S, O'Connell MJ. Gene Signatures in Stage II Colon Cancer: A Clinical Review. Curr Colorectal Cancer Rep 2012;8:225-31. [Crossref] [PubMed]
- Yang YD, Cho H, Koo JY, et al. TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature 2008;455:1210-5. [Crossref] [PubMed]
- Seo Y, Ryu K, Park J, et al. Inhibition of ANO1 by luteolin and its cytotoxicity in human prostate cancer PC-3 cells. PLoS One 2017;12:e0174935 [Crossref] [PubMed]
- Ruiz C, Martins JR, Rudin F, et al. Enhanced expression of ANO1 in head and neck squamous cell carcinoma causes cell migration and correlates with poor prognosis. PLoS One 2012;7:e43265 [Crossref] [PubMed]
- Schroeder BC, Cheng T, Jan YN, et al. Expression cloning of TMEM16A as a calcium-activated chloride channel subunit. Cell 2008;134:1019-29. [Crossref] [PubMed]
- Liu W, Lu M, Liu B, et al. Inhibition of Ca(2+)-activated Cl(-) channel ANO1/TMEM16A expression suppresses tumor growth and invasiveness in human prostate carcinoma. Cancer Lett 2012;326:41-51. [Crossref] [PubMed]
- Diamandis EP, Yousef GM, Soosaipillai AR, et al. Human kallikrein 6 (zyme/protease M/neurosin): a new serum biomarker of ovarian carcinoma. Clin Biochem 2000;33:579-83. [Crossref] [PubMed]
- Lundwall A, Band V, Blaber M, et al. A comprehensive nomenclature for serine proteases with homology to tissue kallikreins. Biol Chem 2006;387:637-41. [Crossref] [PubMed]
- Luo LY, Bunting P, Scorilas A, et al. Human kallikrein 10: a novel tumor marker for ovarian carcinoma? Clin Chim Acta 2001;306:111-8. [Crossref] [PubMed]
- Grum-Schwensen B, Klingelhofer J, Berg CH, et al. Suppression of tumor development and metastasis formation in mice lacking the S100A4(mts1) gene. Cancer Res 2005;65:3772-80. [Crossref] [PubMed]
- Sapkota D, Bruland O, Costea DE, et al. S100A14 regulates the invasive potential of oral squamous cell carcinoma derived cell-lines in vitro by modulating expression of matrix metalloproteinases, MMP1 and MMP9. Eur J Cancer 2011;47:600-10. [Crossref] [PubMed]
- Chen H, Yuan Y, Zhang C, et al. Involvement of S100A14 protein in cell invasion by affecting expression and function of matrix metalloproteinase (MMP)-2 via p53-dependent transcriptional regulation. J Biol Chem 2012;287:17109-19. [Crossref] [PubMed]
- Mashek DG, McKenzie MA, Van Horn CG, et al. Rat long chain acyl-CoA synthetase 5 increases fatty acid uptake and partitioning to cellular triacylglycerol in McArdle-RH7777 cells. J Biol Chem 2006;281:945-50. [Crossref] [PubMed]
- Li LO, Mashek DG, An J, et al. Overexpression of rat long chain acyl-coa synthetase 1 alters fatty acid metabolism in rat primary hepatocytes. J Biol Chem 2006;281:37246-55. [Crossref] [PubMed]
- Gassler N, Roth W, Funke B, et al. Regulation of enterocyte apoptosis by acyl-CoA synthetase 5 splicing. Gastroenterology 2007;133:587-98. [Crossref] [PubMed]
- Matsuyama S, Llopis J, Deveraux QL, et al. Changes in intramitochondrial and cytosolic pH: early events that modulate caspase activation during apoptosis. Nat Cell Biol 2000;2:318-25. [Crossref] [PubMed]
- Fagoonee S, Picco G, Orso F, et al. The RNA-binding protein ESRP1 promotes human colorectal cancer progression. Oncotarget 2017;8:10007-24. [Crossref] [PubMed]
- Shapiro IM, Cheng AW, Flytzanis NC, et al. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet 2011;7:e1002218 [Crossref] [PubMed]
- Ueda J, Matsuda Y, Yamahatsu K, et al. Epithelial splicing regulatory protein 1 is a favorable prognostic factor in pancreatic cancer that attenuates pancreatic metastases. Oncogene 2014;33:4485-95. [Crossref] [PubMed]
- Ishii H, Saitoh M, Sakamoto K, et al. Epithelial splicing regulatory proteins 1 (ESRP1) and 2 (ESRP2) suppress cancer cell motility via different mechanisms. J Biol Chem 2014;289:27386-99. [Crossref] [PubMed]
- Voena C, Varesio LM, Zhang L, et al. Oncogenic ALK regulates EMT in non-small cell lung carcinoma through repression of the epithelial splicing regulatory protein 1. Oncotarget 2016;7:33316-30. [Crossref] [PubMed]
- Yae T, Tsuchihashi K, Ishimoto T, et al. Alternative splicing of CD44 mRNA by ESRP1 enhances lung colonization of metastatic cancer cell. Nat Commun 2012;3:883. [Crossref] [PubMed]
- Marzese DM, Liu M, Huynh JL, et al. Brain metastasis is predetermined in early stages of cutaneous melanoma by CD44v6 expression through epigenetic regulation of the spliceosome. Pigment Cell Melanoma Res 2015;28:82-93. [Crossref] [PubMed]
- Zeng CY, Zhan YS, Huang J, et al. MicroRNA7 suppresses human colon cancer invasion and proliferation by targeting the expression of focal adhesion kinase. Mol Med Rep 2016;13:1297-303. [Crossref] [PubMed]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006;6:857-66. [Crossref] [PubMed]
- Wu DG, Wang YY, Fan LG, et al. MicroRNA-7 regulates glioblastoma cell invasion via targeting focal adhesion kinase expression. Chin Med J (Engl) 2011;124:2616-21. [PubMed]
- Sein TT, Thant AA, Hiraiwa Y, et al. A role for FAK in the Concanavalin A-dependent secretion of matrix metalloproteinase-2 and -9. Oncogene 2000;19:5539-42. [Crossref] [PubMed]
- Chen Y, Du M, Wang J, et al. MiRNA-200a expression is inverse correlation with hepatocyte growth factor expression in stromal fibroblasts and its high expression predicts a good prognosis in patients with non-small cell lung cancer. Oncotarget 2016;7:48432-42. [PubMed]
- Machackova T, Mlcochova H, Stanik M, et al. MiR-429 is linked to metastasis and poor prognosis in renal cell carcinoma by affecting epithelial-mesenchymal transition. Tumour Biol 2016;37:14653-8. [Crossref] [PubMed]
- Zuberi M, Mir R, Das J, et al. Erratum to: Expression of serum miR-200a, miR-200b and miR-200c as candidate biomarkers in epithelial ovarian cancer and their association with clinicopathological features. Clin Transl Oncol 2015;17:840. [Crossref] [PubMed]
- Meng X, Muller V, Milde-Langosch K, et al. Diagnostic and prognostic relevance of circulating exosomal miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer. Oncotarget 2016;7:16923-35. [Crossref] [PubMed]


