
Circular RNA: non-coding RNA with emerging roles in stem cell differentiation
Several different types of non-coding RNA have been shown to contribute to the regulation of pluripotency and renewal of stem cells and thereby participate in embryogenesis and development (1-4). Amongst these, the evidence for microRNA and long non-coding RNA as key players is particularly compelling. Several different individual or clusters of microRNAs have the potential to modulate embryonic stem cell pluripotency and reprogramming. These include microRNAs such as miR-372, miR-290 family, miR-302 family, miR-17-92, miR-106b-25 and miR-106a-363 clusters (5). Similarly, other types of non-coding RNA also have the potential to modulate the transcriptional networks that regulate pluripotency and lineage differentiation (6). The long non-coding RNA lncRNA-ROR, for example, can play a role in reprogramming of fibroblast into induced pluripotent stem cells (iPSCs) (7). LncRNA ROR can bind to pluripotency-associated transcription factors (PATFs) such as OCT4, SOX2 and NANOG that contribute to the maintenance of pluripotency in human ESC, as well as act as a sponge for miR-145, a microRNA that is implicated in the repression of genes involved in pluripotency (8).
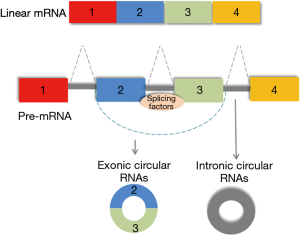
Recent studies have implicated yet another type of non-coding RNA, the circular RNAs, in the control of pluripotency in humans. Circular RNAs are a class of non-coding RNA, so called because they differ from linear RNAs and assume a closed conformation. These RNAs form covalently closed loops generated from back splicing of exons or introns (or both) (Figure 1). The 3' and the 5' ends join together resulting in the formation of a continuous loop, and a conformation that renders them resistant to degradation by exonucleases. The potential contribution of circular RNA to regulation of gene expression is being recognized (9). A functional role of circular RNAs in controlling pluripotency in hESC was described in a study by Yu et al. (10). This study identified circular RNAs that are enriched in undifferentiated human ESCs and iPSCs. Two of these circular RNAs, circBIRC6 and circCORO1C were noted to be functionally related to maintenance of pluripotency. Modulation of circular RNAs using small hairpin RNA to specifically target circular junctions resulted in cellular differentiation and loss of pluripotency. This was associated with downregulated expression of PATFs such as NANOG, OCT4, KLF4 and MYC and a corresponding increase in the expression of lineage-related transcription factors (LRTF) Brachyury, SOX17 and SOX1.
Enforced expression of the three circular RNAs in hESCs using minigene constructs, in the absence of alterations in expression of the un-spliced circular RNA precursors, did not result in a loss of pluripotency. In differentiating hESCs, ectopic expression of circBIRC6 or circCORO1C but not circMAN1A2 increased the expression of PATFs and decreased LRTFs. Thus, circBIRC6 or circCORO1C contribute to maintenance of pluripotency in differentiated hESCs. At the same time, expression of neither circBIRC6 or circCORO1C was sufficient to reprogram somatic cells into iPSCs. However, the co-expression of circular RNAs and PATFs improved TF-mediated reprogramming in somatic fibroblasts.
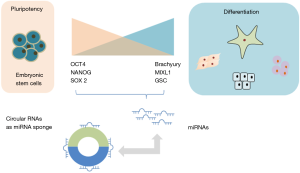
The affinity of binding of miRNAs with circular RNAs is reported to be greater than that of binding to their endogenous target mRNA. Thus, circular RNA can act as a sponge to sequester miRNA and reduce their effects on endogenous targets. For example, ciRS-7 can sequester miR7 thereby interfering with miR7 effect on the suppression of epithelial to mesenchymal transition and cancer progression. Circular RNAs have also been reported to act as alternative splicing regulators or transcription factors (11). CircBIRC6 but not circCORO1C was enriched in AGO immunoprecipitates suggesting that circBIRC6 could possess miRNA related functions. Indeed, circBIRC6 interacted with miR-34a and miR-145 in ESCs, acting as a sponge to maintain pluripotency (Figure 2). Both of these microRNAs are associated with pluripotency associated genes and can restrain somatic reprogramming by repressing the expression of these genes (3,4). Targeting sites for both of these miRNAs are also present on circBIRC6. A decreased expression of these miRNA was noted in undifferentiated hESCs but not in differentiated hESCs.
At first, eukaryotic circular RNAs were thought to represent artefacts in RNA sequencing. It is now recognized that these RNA molecules can be generated as a result of alternative splicing processes. Splicing results in the pairing of exons in a sequential order leading to linear RNA formation, but can also result in new transcripts such as circular RNA through back-splicing. Understanding the mechanisms of generation of circular RNAs, and the settings in which these processes occur will be necessary in elucidating their functional roles. Yu et al. also reported the involvement of splicing factors in generating circular RNA in human ESCs. The splicing factor ESRP1 was shown to be upregulated in ESCs, and involved in the formation of circBIRC6. Moreover, NANOG and Oct4 were shown to regulate ESRP1 expression. Further studies to extend this work may involve the generation and study of conditional knockouts of key proteins involved in splicing.
Improved techniques for the detection of circular RNA have resulted in characterization of their prevalence, expression in health and disease states, and their potential functional roles. The detection of circular RNA molecules has been limited because of their low abundance and the lack of robust methods for their detection. However, the advent of gene sequencing technologies, and the use of specific analytical pipelines for the detection of circular RNA have facilitated the recognition and identification of these unique non-coding RNA. Widespread expression of circular RNAs have been observed, often in a cell type, tissue type or context-specific setting. Emerging data show that circular RNAs are stable, abundant and conserved among various biological systems. These studies are enhancing our knowledge of the role of these RNAs. Understanding the contributions of circular RNAs in the regulation of gene expression and control of pluripotency has relevance for other disease areas. For example, circular RNAs have been implicated in cancer where they may participate in tumorigenesis and disease progression (9). Pluripotent stem cells share many features with tumor cells, such as self-renewal and proliferation, while some of the transcriptional regulatory mechanisms and transcription factors involved in pluripotency may also participate in regulating tumorigenesis.
Understanding the processes that control the formation of circular RNA in different settings will be central to understanding their function. More studies are needed to elucidate the mechanisms by which a specific gene can result in various distinct transcripts that may have very different functional roles. We are only now beginning to recognize the incredible complexity of the human genome.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Chunlin Ou, MD, PhD (Cancer Research Institute of Central South University, Changsha, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.12.26). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet 2011;12:136-49. [Crossref] [PubMed]
- Wright JE, Ciosk R. RNA-based regulation of pluripotency. Trends Genet 2013;29:99-107. [Crossref] [PubMed]
- Choi YJ, Lin CP, Ho JJ, et al. miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat Cell Biol 2011;13:1353-60. [Crossref] [PubMed]
- Xu N, Papagiannakopoulos T, Pan G, et al. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 2009;137:647-58. [Crossref] [PubMed]
- Li N, Long B, Han W, et al. microRNAs: important regulators of stem cells. Stem Cell Res Ther 2017;8:110. [Crossref] [PubMed]
- Ghosal S, Das S, Chakrabarti J. Long noncoding RNAs: new players in the molecular mechanism for maintenance and differentiation of pluripotent stem cells. Stem Cells Dev 2013;22:2240-53. [Crossref] [PubMed]
- Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J 2012;31:522-33. [Crossref] [PubMed]
- Pan Y, Li C, Chen J, Zhang K, et al. The Emerging Roles of Long Noncoding RNA ROR (lincRNA-ROR) and its Possible Mechanisms in Human Cancers. Cell Physiol Biochem 2016;40:219-29. [Crossref] [PubMed]
- Greene J, Baird AM, Brady L, et al. Circular RNAs: Biogenesis, Function and Role in Human Diseases. Front Mol Biosci 2017;4:38. [Crossref] [PubMed]
- Yu CY, Li TC, Wu YY, et al. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat Commun 2017;8:1149. [Crossref] [PubMed]
- Xin Z, Ma Q, Ren S, et al. The understanding of circular RNAs as special triggers in carcinogenesis. Brief Funct Genomics 2017;16:80-6. [PubMed]


