
Prostate cancer: unmet clinical needs and RAD9 as a candidate biomarker for patient management
Introduction
Prostate cancer is the most prevalent form of non-cutaneous cancer in men, and a major cause of mortality and morbidity [(1) www.cancer.org/cancer/prostate-cancer/about/key-statistics.html]. It is estimated by the American Cancer Society that for the year 2018, there will be 164,690 new cases of prostate cancer diagnosed in the United States, and a staggering 29,430 of these men will die because of the disease. About 1 in 7 men will be diagnosed with prostate cancer in their lifetimes, and about 1 out of every 39 men will die because of it, so there is a clear need for biomarkers to enhance diagnostic power and inform treatment decision. Well-established, minimally invasive clinical tests, such as monitoring prostate-specific antigen (PSA) levels in blood, and digital rectal exams (DRE), allow screening and tracking of disease progression. If results are deemed suspicious, follow up biopsy and histological grading is performed to establish the presence and characteristics of the cancer. Initial treatment for localized disease may involve an active surveillance approach, partial or radical prostatectomy, and/or radiotherapy applied as an external beam or radioactive seed implant. More advanced disease treatment entails androgen deprivation therapy, chemotherapy, or immunotherapy to boost the immune system. Treatment failure is usually caused by lack of local control and aggressive metastasis of tumor cells to distal sites, notably bone (2). Understanding the underlying genetic basis of prostate cancer would allow the development of novel biomarkers and unique targets for therapy from an informed, mechanistic perspective.
Molecular diagnostic tools for prostate cancer: current non-invasive tests
Prostate cancer is a highly curable disease if detected early. Although there has been considerable progress in the development of novel therapeutic interventions, the introduction of effective diagnostic biomarkers has lagged behind. Therefore, an important clinical need is the establishment of better diagnostic tools, preferably non-invasive or minimally invasive, that would permit accurate detection and assessment of prostate cancer, especially in terms of stratifying risk for disease progression in men confirmed by biopsy to have this cancer. Current diagnostic procedures are valuable yet have deficiencies. Although blood PSA quantification is widely used and is considered a valuable diagnostic tool, a variety of conditions or activities other than prostate cancer can elevate levels of this protein, such as benign prostatic hyperplasia (BPH), prostatitis, sexual activity, and exercise (3). Further, it is not clear if blood PSA levels between 2 and 10 ng/mL for men older than 50 years unequivocally indicate cancer (American Cancer Society), and this can lead to painful, often unnecessary biopsy. About 15% of men with a PSA below 4 will have prostate cancer indicated on biopsy, and for those between PSA 4 and 10 ng/mL there is an ~25% chance of cancer. Even for men with PSA above 10 ng/mL, the chance of having prostate cancer is just over 50%, not 100%. Moreover, aggressive neuroendocrine prostate cancer has little or no PSA (4). There are also some gray areas in terms of characterizing prostate tissue and significance with respect to disease. Well selected patients with tumors scored as Gleason 6 low volume disease are usually followed by active surveillance, yet a fraction of those men, who cannot be identified beforehand, eventually require immediate, aggressive treatment. In the setting of post-prostatectomy or radiation treatment, advances such as the development of ultrasensitive PSA tests and the application of the Phoenix definition of biochemical failure after radiation treatment have improved the ability to monitor patients. However, there is still a great unmet need for similar advances to assess disease in patients at the pre-treatment stage. Thus, patients would benefit by improved and, in particular, non-invasive predictive biomarkers that could indicate presence of cancer or risk of developing advanced disease once cancer is identified.
Total PSA has satisfactory sensitivity but poor specificity as a tumor marker for prostate cancer. It has several drawbacks, and a number of markers have become available in the last few years to aid PSA in the detection of prostate cancer. These include PCA3, prostate health index (phi), and 4K score. Each marker/test has shown improvement in performance characteristics over total PSA alone, but each has limitations and none of the markers are ideal. There are differences in sample requirements, site of testing (routine laboratory vs. dedicated reference laboratory), test performance, and intended use. Thus, a perfect marker for prostate cancer detection useful in risk stratification, and that can be obtained non-invasively, still remains elusive. A number of studies have been conducted to characterize these performance features better, and results show that sensitivity as well as specificity can be improved further for each of the assays. Examples are described below.
PCA3 is a highly over-abundant RNA in prostate cancers, with cancer cells expressing 60–100× greater levels than normal cells. The test quantifies PCA3 versus PSA RNA and has been commercialized as Progensa PCA3 assay by Gen-probe Inc. It requires a post-DRE urine sample. The final result is reported as a “PCA3 score” and is calculated as a ratio of (PCA3 RNA/PSA RNA) ×1,000. Testing requires a doctor’s visit, as specimens must be collected as a post-DRE sample in a urologist’s office and subsequently shipped to the testing site. Additional details are available at www.gen-probe.com. PCA3 has a pooled sensitivity of 65% and specificity of 73%, based on a meta-analysis of 46 clinical trials (5).
The phi is commercialized by Beckman Coulter Inc. and is an U.S. Food & Drug Administration (FDA) approved blood test used as an aid in distinguishing prostate cancer from benign prostatic conditions [www.beckmancoulter.com; (6)]. It is a calculation based on an algorithm combining total PSA, free PSA, and another form of PSA, namely [-2]proPSA. The test measures these analytes in blood and can be performed on standard clinical analyzers available through Beckman Coulter Inc. Thus, it provides an opportunity for most clinical labs to offer this test as a panel. However, one disadvantage is that the test kit is only offered as a bundle, and all three tests must be performed on Beckman instrumentation. Total PSA, if quantified on other analyzers, cannot be combined with the remaining analytes, so clinical labs would need to offer this test panel in addition to any other PSA testing they’ve already performed. The phi test outperforms PSA alone (7) and has 90% clinical sensitivity and 31% specificity at a phi value of 27.0 (www.beckmancoulter.com), allowing for a reduction of nearly 1/3 of all biopsies while detecting 90% of cancers.
A comparison of the PSA, PCA3, and phi methods in terms of area under the curve (AUC) and the sensitivity at 85% specificity (85% probability that a man with PSA <3.0 does not have cancer) is given in Table 1.
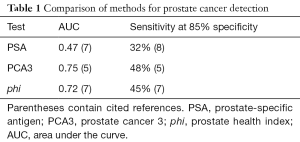
Full table
The 4K score combines the levels of four prostate-specific kallikreins as well as clinical information into an algorithm that predicts disease aggressiveness. Using such a test, a low risk patient can be placed on an active surveillance regimen, whereas a high-risk individual can elect to undergo biopsy for further evaluation. The test includes the measurement of four kallikrein proteins in blood. These include free and intact PSA, hk2, and hk3. This test is only available through BioReference Laboratories, a division of OPKO Health Inc., so samples must be submitted to them for analysis. Overall, this test does very well (AUC =0.82) in identifying men with clinically significant disease, Gleason ≥7 (9).
In addition to the above, other commercial tests have become available that employ classifiers measuring analytes present in tissue samples. These tests require invasively obtained samples (human prostate tissue, either biopsy or resection material) and cannot be readily implemented in clinical laboratories, as samples need to be shipped to a single commercial laboratory.
One example of such a diagnostic is the Oncotype Dx Prostate Cancer test from Genomic Health (www.genomichealth.com). It is for men with newly diagnosed, early stage prostate cancer and is used to determine the need for treatment. Such a test is useful as only ~3% of such individuals will die of a prostate cancer-specific death. The test utilizes biopsy material and is a multi-gene classifier that predicts aggressive disease at the time of diagnosis. In such a manner, it can be used to individualize treatment for patients.
A second example is Prolaris (www.prolaris.com), also used to predict disease aggressiveness based on analysis of biopsy tissue. This test specifically assesses genes involved in cellular proliferation and can be used to predict disease-specific mortality. In an additional application, this test can also be used to predict biochemical recurrence in post-prostatectomy patients. When employed for this purpose, the gene signature becomes a useful risk stratification tool that can assist in defining treatment strategy for patients after surgery.
The stratification of biopsy-confirmed prostate cancer patients for risk of progression to metastatic disease is a critical issue that needs to be addressed, in terms of informing decisions to manage these men with less invasive therapy such as active surveillance, or to consider more aggressive treatment. Gnanapragasam and coworkers (10) recently developed a 5-stage risk stratification system based not just on surrogate markers of the disease, such as biochemical recurrence, but prostate cancer-specific mortality. Their system has excellent predictive power and takes into consideration PSA level at diagnosis, clinical T stage, and the novel grouping system developed by the International Society of Urological Pathology (ISUP), which is based on glandular architecture (11).
Ideally, regardless of type of patient specimen being analyzed, predictive biomarkers should be identifiable and even useful in some instances as targets for therapy. A major impediment to this precision medicine approach is the heterogeneity of cancer-characteristic molecular profiles, making the identification of tumors and discrimination of their differential behaviors very difficult (12). To complicate the search for ideal signatures, prostate cancer is often multifocal with clonal subpopulations bearing varied histological and molecular abnormalities. This heterogeneity exists not only within but also between patients (13,14). Nevertheless, despite these challenges, much research is focused on the further identification of better predictive biomarkers that are more sensitive and specific than what is currently available and in clinical use. For example, Irshad et al. (15) reported that the three-gene set of FGFR1, PMP22, and CDKN1A tested in prostate biopsies together accurately predicted outcome of low Gleason score prostate tumors. Lalonde et al. (16) refined a 100-gene set down to 31 in prostate tissue that can identify prostate cancer patients with high biochemical recurrence rates [hazard ratio (HR) =2.73, P<0.001] and those who eventually develop metastasis (HR =7.79, P<0.001). When combined with more standard tests, predictive value increases further.
Although there has been great success in the identification of novel biomarkers, only a few of them have advanced to clinical practice. Of those that are in use today, their performance characteristics demonstrate that there is still room for improvement. For others that are earlier in the translational pipeline, they also are not ideal—some require sample material that is invasively obtained, and for most others, further work is still required for validation, translation to clinical assays, and ultimately to demonstrate their utility. Thus, there is a pressing need for a biomarker that can be translated to a clinical assay useful in the management of prostate cancer patients. We present below one such class of proteins, that of the DNA damage response, and then propose RAD9, a member of that group, as a candidate biomarker for prostate cancer.
DNA damage response proteins as predictive cancer biomarkers and anti-cancer targets
DNA repair proteins and other DNA damage response factors have been evaluated as prognostic biomarkers and their status has been included in strategies for cancer treatment [e.g., use of poly-(ADP-ribose)-polymerase (PARP) inhibitors for BRCA1 mutant tumors; (17-19)]. Chemotherapy and radiotherapy primarily kill tumor cells by damaging DNA, and thus tumor DNA repair capacity should predictably, directly affect therapeutic efficacy and prognostic outlook. In addition, DNA repair should be manipulatable to optimize tumor cell killing.
Genetically heritable human DNA damage response syndromes are infrequent but exemplify the importance of DNA repair related mechanisms with respect to carcinogenesis and response to agents used for cancer therapy. Examples of such heritable syndromes include ataxia telangiectasia-mutated [DNA damage response signaling pathway deficiency; (20)], breast cancer associated 1 and 2 [interstrand crosslink and double-strand DNA break repair deficiency; (21)], Lynch syndrome [also known as hereditary non-polyposis colorectal cancer; mismatch repair deficiency; (22)], Nijmegen breakage syndrome [defective in sensing DNA double strand breaks; (23)], and Werner syndrome [DNA double strand break repair deficiency; (24)]. Generally, individuals afflicted with these disorders are at high risk for developing cancer, and their cells are genetically unstable as well as hypersensitive to the killing effects of DNA damaging agents.
In terms of radiotherapy efficacy for cancer patients with germline DNA repair gene alterations, the results are mixed. Fourquet et al. (25) found that inherent BRCA1 and BRCA2 mutations were associated with better clinical response of breast cancer patients to radiotherapy and higher breast conservation rates, relative to patients not bearing the genetic alterations. Further, BRCA1 mutation was the sole predictor of successful breast conservation. In contrast, germline BRCA1 or BRCA2 mutations in prostate cancer patients are associated with poor survival outcome after radiotherapy (26,27). Zanusso et al. (28) found that specific germline polymorphisms in ERCC1 and EXO1 are significantly associated with biochemical recurrence of prostate cancer in patients treated with radiotherapy, and a polymorphism in MSH6 is associated with worse overall survival. However, decision curve analysis and sensitivity analysis did not provide conclusive guidance with respect to clinical impact of the polymorphisms identified. The reasons for these different, sometimes counterintuitive therapeutic responses in the context of germline DNA repair gene alterations is not clear, and need additional investigation.
Ideally for clinical exploitation, instead of germline mutations as per the heritable disorders or DNA repair gene polymorphisms mentioned, somatic cancer cell genetic alterations have been observed that manifest as differential repair capacity between tumor and surrounding tissues (29-31). There are numerous mechanisms for repairing damaged DNA, and there is overlap with respect to pathways that can process the same type of damage (18). For example, DNA double strand breaks in cells can be repaired by homologous recombination in the S and G2 cell cycle phases, or by non-homologous end joining in all phases of the cell cycle. Moreover, DNA base lesions are repaired by base excision repair (BER). However, defective BER can produce DNA single strand breaks, which can be converted to double strand breaks and repaired predominantly by homologous recombination in S and G2. Tumor cells relative to non-cancer cells are often defective in DNA damage response pathways, most notably homologous recombination repair due to prevalence of BRCA1 or BRCA2 mutations (19,29). In these instances, tumor cells rely on backup mechanisms, such as BER, and are thus comparatively more vulnerable to DNA damage.
PARP is a family of enzymes that participate in repair of DNA damage, by several processes including BER (32), homologous recombination repair (33), non-homologous end joining (34), and alternative non-homologous end joining (35). Notably, PARP-1 and PARP-2 senses single strand nicks in DNA, then cause extensive ADP-ribosylation, and that results in the recruitment of XRCC1, DNA ligase III, and DNA polymerase beta to the damaged site, which then initiates BER. PARP inhibitors have shown promise in terms of causing synthetic lethality in genitourinary malignancies with BRCA1 or BRCA2 deficiencies (36), or as radiosensitizers to enhance radiotherapy (37). Niraparib, olaparib, and rucaparib each inhibit both PARP-1 and PARP-2, and have been FDA approved for use by women with advanced ovarian cancer (38). Niraparib is approved for use regardless of whether or not women have germline or inherited, BRCA1 or BRCA2 gene mutations. Olaparib can only be used by women with deleterious germline BRCA1 or BRCA2 mutations, and rucaparib is approved for use by those with germline or somatic BRCA mutations.
RAD9 and prostate cancer
As mentioned earlier, current tests for prostate cancer are not ideal, and there is an urgent need for improved markers. RAD9 has the potential to serve in that capacity. This protein functions as an oncogene in prostate cancer. RAD9 level is abnormally elevated in multiple prostate cancer cell lines (14−22 fold, relative to non-cancer prostate cells). Hypermethylation of a transcription suppressor region in RAD9 intron 2, or gene amplification, are two mechanisms responsible (39). Interestingly, these mechanisms are also responsible for the aberrantly high expression of RAD9 in some breast tumors (40). Decreased RAD9 abundance mediated by RNA interference in prostate cancer cell lines dramatically reduces tumorigenicity in nude mouse xenographs, indicating that RAD9 has a critical, causal role in this type of cancer (39). More than 45% of prostate tumors have aberrantly high levels of RAD9. There is a strong, significant (P value <0.001) positive association between frequency and intensity of immunohistochemical staining for RAD9 protein and cancer stage in human prostate tumor specimens, where the highest protein levels are in advanced Stage III and IV tumors as well as metastases. Further, RAD9 functions in tumor metastasis, and thus its activity is not limited to initial tumor formation. Down regulation of RAD9 in prostate cancer cells in culture impairs metastasis-related phenotypes, including migration, invasion, and anchorage-independent growth, as well as sensitizes those cells to anoikis (41,42), which is programmed death induced when cells are not attached to a matrix (43). Typically, metastatic cells are anoikis resistant, enabling them to remain viable as they travel through the bloodstream for eventual localization in distal sites. RAD9 affects cell migration and invasion by modulating integrin β1 protein level, and anoikis resistance by modulating AKT kinase activation (42).
The activity of androgen receptor is critical for prostate maintenance, as well as for prostate cancer and treatment of patients with the late stage of the disease. RAD9 is intimately involved in androgen receptor function and is itself androgen responsive. Wang and colleagues (44) reported that RAD9 acts as a negative co-regulator of the transactivation function of androgen receptor. A motif within the C-terminal region of RAD9 binds androgen receptor, and thus interrupts androgen-induced androgen receptor N-terminus and C-terminus interactions, repressing the transactivation function of the receptor that normally controls downstream effector gene expression, such as that encoding prostate specific antigen (45). Moehren et al. (46) demonstrated that androgen can induce RAD9 expression 14.1-fold through androgen receptor binding to promoter-localized androgen receptor elements. However, human prostate cancer DU145 and PC-3 cells have high levels of RAD9 and reduction by RNA interference reduces or eliminates the tumorigenic potential of those cells when injected subcutaneously into nude mice (39), although both cell lines are androgen receptor negative (47). Therefore, the biological significance of the relationship between RAD9 and androgen receptor with respect to prostate carcinogenesis is not straightforward and needs to be evaluated further.
RAD9 molecular networks
RAD9, the DNA damage response, and DNA repair
RAD9 has many functions that control genomic stability, which is a hallmark driving force for cancer development and progression, so it is not surprising that the protein is critical for prostate carcinogenesis and might be useful as a cancer biomarker or treatment target. The human protein can form and function as part of a heterotrimeric complex with HUS1 and RAD1 [i.e., the 9-1-1 complex (48-51)], but also act independently (41,52). Using Rad9 knockout mice, as well as human and mouse cells with varied RAD9 status, it was demonstrated that loss of RAD9 causes extreme cellular sensitivity to a wide variety of radiations and chemicals that damage DNA, and reduces genomic stability by numerous activities. RAD9 is essential for maintaining genomic integrity even in the absence of exogenous DNA damaging agents in addition, as loss of function causes increased frequencies of spontaneous chromosome and chromatid breaks, gene mutation, and micronuclei (53,54). RAD9 plays a number of key roles that promote resistance to and removal of DNA damage.
RAD9 regulates cell cycle checkpoints, which transiently delay cell cycle progression at specific junctures to provide extra time for restoration of DNA integrity before entry into critical cell cycle phases, an event that could be lethal in the presence of DNA damage (53,55,56). Cell cycle checkpoint control is a highly complex process, which is initiated when DNA damage occurs. ATRIP physically interacts with RPA that is bound to DNA, and this stimulates the RAD17-RFC2-5 complex to recruit 9-1-1 to the damaged site (57). This then leads to recruitment of TOPBP1 to the site by an interaction with the phosphorylated C-terminal tail of RAD9. TOPBP1 then activates ATR, which depends upon ATRIP and is facilitated by RHINO (58,59). CHK1 and other downstream target proteins are then phosphorylated and activated. The C-terminus of RAD9 also helps bring CLASPIN to the site of DNA damage, and this event additionally aids CHK1 activation (60,61). This signaling cascade results in cell cycle arrest, as well as replication fork stabilization and restart as needed (62).
RAD9 also plays important roles in at least five DNA repair pathways. RAD9 contributes to homologous recombination repair through interaction with RAD51 (63). RAD9 has more than one activity in BER, either by binding and modulating the activity of other proteins critical for the process, or in one instance, regulating the level of a key protein, NEIL1, by direct transcriptional control of the corresponding gene (64-68). RAD9 also participates in nucleotide excision repair by maintaining DDB2 protein level (69), mismatch repair by physical interaction with MLH1 (70), and alternative non-homologous end joining (71), such as microhomology-mediated DNA strand annealing by promoting end joining of non-repetitive sequences through the activity of DNA pol λ and by limiting triplet expansion via DNA pol β (72).
Apoptosis
Aside from roles in cell cycle checkpoint control and DNA repair, RAD9 functions in programmed cell death as a pro-apoptotic (73,74) or anti-apoptotic protein (54), depending upon cell context and level of abundance. The N-terminal region of RAD9 contains a BH3-like domain that binds anti-apoptotic proteins BCL-XL and BCL-2 to promote programmed cell death. Phosphorylation regulates the binding. c-ABL can phosphorylate tyrosine-28 located in the BH3-like domain of RAD9, and that enhances BCL-XL binding and leads to apoptosis (75). Phosphorylation of RAD9 by protein kinase Cδ (PKCδ) modulates the interaction between RAD9 and BCL-2, and the resulting apoptotic response to DNA damage (76).
RAD9 functions as a gene-specific transcription factor
Many known cellular roles of RAD9 are mediated by protein-protein interactions whereby RAD9 through physical binding alters binding partner activity. However, RAD9 also exerts control of cellular functions by regulating transcription of select target genes (77). RAD9 controls expression of cell cycle/senescence gene CDKN1A (p21waf1/Cip1) (52,78) and BER gene NEIL1 (68). For these genes, RAD9 binds established transcription factor p53 binding consensus sequences near respective gene promoters (79-81). Interestingly, like p53, RAD9 also represses transcription. For example, Wen et al. (82) reported that RAD9 can suppress epithelial-mesenchymal transition (EMT) through inhibition of SLUG transcription, but this can be cancer-type specific. A limited microarray gene expression study indicated 92 genes are regulated by RAD9 (52). Whether they are directly or indirectly controlled by RAD9, and the significance of p53 binding consensus sequences, have yet to be determined.
RAD9 as a precision medicine tool for the diagnosis and treatment of prostate cancer
Given the important functions of RAD9 in maintaining genomic integrity through multiple mechanisms, and the demonstrated specific roles of RAD9 in prostate tumorigenesis and metastasis, the protein should be a good candidate for the development of precision cancer diagnostic and therapeutic tools. Zhu et al. (39) already showed that RAD9 is detected in prostate cancer tissues. It would be ideal if the protein could be monitored in a non-invasive fashion as well, for example directly in clinical body fluids or in isolated blood or urine vesicles, such as exosomes, where live cells package and shed intracellular components by exocytosis. Exosomes primarily contain proteins usually localized in cell cytosolic or endosomal compartments (83), and RAD9 is found in the cytoplasm as well as in the nucleus (74,84,85). However, detection of RAD9 in body fluids or specifically in exosomes has not yet been demonstrated. In addition, with respect to being a direct therapeutic target, the pleiotropic effects of RAD9 inhibition might be problematic when blocking activity and thereby cause unwanted deleterious effects. In that regard, defining and targeting downstream effectors for inhibition might provide a more specific, precise mechanism to eradicate tumors. This has potential, as RAD9 is a transcription factor with a specific set of regulated target genes.
Conclusions
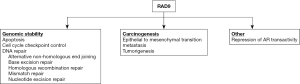
Prostate cancer is highly prevalent among men, and there is heterogeneity with respect to initial presentation, prognosis, and underlying mechanism that contributes to the disease. A better understanding of critical molecular details of the major driving processes would allow development of precision diagnostic and therapeutic tools to reduce morbidity and mortality. Although several non-invasive clinical tests have become available in recent years, none are ideal and thus there is still a need for an optimal biomarker that can be translated for use in a clinical setting. RAD9 can potentially fill this current void, certainly in biopsy material, but additional work is required to demonstrate that the protein can be detected non-invasively. RAD9 controls a number of cellular activities through multiple processes that affect the disease. Figure 1 summarizes those activities. Roles in maintaining genomic integrity are considered fundamental determinants of cancer initiation and progression. However, the function of RAD9 as a transcription activator or repressor, as well as the relationship with androgen receptor and responsiveness to testosterone, suggest additional, more specific but complex functions of the protein in prostate cancer. Evaluation of the activities of RAD9 in this cancer, and the identification of downstream as well as regulatory upstream elements that control RAD9 abundance should provide important new biomarkers for disease diagnosis and likely also unique therapeutic targets.
Acknowledgments
Funding: This work was supported by the National Institutes of Health (grant R01CA130536).
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Israel Deutsch, James McKiernan, Charles Drake) for the series “Prostate Cancer: Current Understanding and Future Directions” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.01.21). The series “Prostate Cancer: Current Understanding and Future Directions” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Shen MM, Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev 2010;24:1967-2000. [Crossref] [PubMed]
- Nandana S, Chung LW. Prostate cancer progression and metastasis: potential regulatory pathways for therapeutic targeting. Am J Clin Exp Urol 2014;2:92-101. [PubMed]
- Tarhan F, Orçun A, Küçükercan I, et al. Effect of prostatic massage on serum complexed prostate-specific antigen levels. Urology 2005;66:1234-8. [Crossref] [PubMed]
- Hu CD, Choo R, Huang J. Neuroendocrine differentiation in prostate cancer: a mechanism of radioresistance and treatment failure. Front Oncol 2015;5:90. [Crossref] [PubMed]
- Cui Y, Cao W, Li Q, et al. Evaluation of prostate cancer antigen 3 for detecting prostate cancer: a systematic review and meta-analysis. Sci Rep 2016;6:25776. [Crossref] [PubMed]
- Loeb S, Sanda MG, Broyles DL, et al. The prostate health index selectively identifies clinically significant prostate cancer. J Urol 2015;193:1163-9. [Crossref] [PubMed]
- Tosoian JJ, Druskin SC, Andreas D, et al. Use of the Prostate Health Index for detection of prostate cancer: results from a large academic practice. Prostate Cancer Prostatic Dis 2017;20:228-33. [Crossref] [PubMed]
- Meigs JB, Barry MJ, Oesterling JE, et al. Interpreting results of prostate-specific antigen testing for early detection of prostate cancer. J Gen Intern Med 1996;11:505-12. [Crossref] [PubMed]
- Parekh DJ, Punnen S, Sjoberg DD, et al. A multi-institutional prospective trial in the USA confirms that the 4Kscore accurately identifies men with high-grade prostate cancer. Eur Urol 2015;68:464-70. [Crossref] [PubMed]
- Gnanapragasam VJ, Lophatananon A, Wright KA, et al. Improving Clinical Risk Stratification at Diagnosis in Primary Prostate Cancer: A Prognostic Modelling Study. PLoS Med 2016;13:e1002063 [Crossref] [PubMed]
- Patel KM, Gnanapragasam VJ. Novel concepts for risk stratification in prostate cancer. J Clin Urol 2016;9:18-23. [Crossref] [PubMed]
- Oliveira-Barros EG, Nicolau-Neto P, Da Costa NM, et al. Prostate cancer molecular profiling: the Achilles heel for the implementation of precision medicine. Cell Biol Int 2017;41:1239-45. [Crossref] [PubMed]
- Fraser M, Berlin A, Bristow RG, et al. Genomic, pathological, and clinical heterogeneity as drivers of personalized medicine in prostate cancer. Urol Oncol 2015;33:85-94. [Crossref] [PubMed]
- Boutros PC, Fraser M, Harding NJ, et al. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat Genet 2015;47:736-45. [Crossref] [PubMed]
- Irshad S, Bansal M, Castillo-Martin M, et al. A molecular signature predictive of indolent prostate cancer. Sci Transl Med 2013;5:202ra122 [Crossref] [PubMed]
- Lalonde E, Alkallas R, Chua MLK, et al. Translating a Prognostic DNA Genomic Classifier into the Clinic: Retrospective Validation in 563 Localized Prostate Tumors. Eur Urol 2017;72:22-31. [Crossref] [PubMed]
- Lieberman HB. DNA damage repair and response proteins as targets for cancer therapy. Curr Med Chem 2008;15:360-7. [Crossref] [PubMed]
- Bhattacharya S, Asaithamby A. Repurposing DNA repair factors to eradicate tumor cells upon radiotherapy. Translational Cancer Res 2017;6:S822-39. [Crossref]
- Mahamud O, So J, Chua MLK, et al. Targeting DNA repair for precision radiotherapy: Balancing the therapeutic ratio. Curr Probl Cancer 2017;41:265-72. [Crossref] [PubMed]
- Rothblum-Oviatt C, Wright J, Lefton-Greif MA, et al. Ataxia telangiectasia: a review. Orphanet J Rare Dis 2016;11:159. [Crossref] [PubMed]
- Stecklein SR, Jensen RA. Identifying and exploiting defects in the Fanconi anemia/BRCA pathway in oncology. Transl Res 2012;160:178-97. [Crossref] [PubMed]
- Martín-López JV, Fishel R. The mechanism of mismatch repair and the functional analysis of mismatch repair defects in Lynch syndrome. Fam Cancer 2013;12:159-68. [Crossref] [PubMed]
- Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nat Rev Mol Cell Biol 2011;12:90-103. [Crossref] [PubMed]
- Brosh RM Jr, Bohr VA. Human premature aging, DNA repair and RecQ helicases. Nucleic Acids Res 2007;35:7527-44. [Crossref] [PubMed]
- Fourquet A, Stoppa-Lyonnet D, Kirova YM, et al. Familial breast cancer: clinical response to induction chemotherapy or radiotherapy related to BRCA1/2 mutations status. Am J Clin Oncol 2009;32:127-31. [Crossref] [PubMed]
- Castro E, Goh C, Olmos D, et al. Germline BRCA mutations are associated with higher risk of nodal involvement, distant metastasis, and poor survival outcomes in prostate cancer. J Clin Oncol 2013;31:1748-57. [Crossref] [PubMed]
- Castro E, Goh C, Leongamornlert D, et al. Effect of BRCA Mutations on Metastatic Relapse and Cause-specific Survival After Radical Treatment for Localised Prostate Cancer. Eur Urol 2015;68:186-93. [Crossref] [PubMed]
- Zanusso C, Bortolus R, Dreussi E, et al. Impact of DNA repair gene polymorphisms on the risk of biochemical recurrence after radiotherapy and overall survival in prostate cancer. Oncotarget 2017;8:22863-75. [Crossref] [PubMed]
- Dhawan M, Ryan CJ, Ashworth A. DNA Repair Deficiency Is Common in Advanced Prostate Cancer: New Therapeutic Opportunities. Oncologist 2016;21:940-5. [Crossref] [PubMed]
- Rimar KJ, Tran PT, Matulewicz RS, et al. The emerging role of homologous recombination repair and PARP inhibitors in genitourinary malignancies. Cancer 2017;123:1912-24. [Crossref] [PubMed]
- Teply BA, Antonarakis ES. Treatment strategies for DNA repair-deficient prostate cancer. Expert Rev Clin Pharmacol 2017;10:889-98. [Crossref] [PubMed]
- Li M, Yu X. The role of poly(ADP-ribosyl)ation in DNA damage response and cancer chemotherapy. Oncogene 2015;34:3349-56. [Crossref] [PubMed]
- Schultz N, Lopez E, Saleh-Gohari N, et al. Poly(ADP-ribose) polymerase (PARP-1) has a controlling role in homologous recombination. Nucleic Acids Res 2003;31:4959-64. [Crossref] [PubMed]
- Mansour WY, Rhein T, Dahm-Daphi J. The alternative end-joining pathway for repair of DNA double-strand breaks requires PARP1 but is not dependent upon microhomologies. Nucleic Acids Res 2010;38:6065-77. [Crossref] [PubMed]
- Cheng Q, Barboule N, Frit P, et al. Ku counteracts mobilization of PARP1 and MRN in chromatin damaged with DNA double-strand breaks. Nucleic Acids Res 2011;39:9605-19. [Crossref] [PubMed]
- O’Kane GM, Connor AA, Gallinger S. Characterization, Detection, and Treatment Approaches for Homologous Recombination Deficiency in Cancer. Trends Mol Med 2017;23:1121-37. [Crossref] [PubMed]
- Lesueur P, Chevalier F, Austry JB, et al. Poly-(ADP-ribose)-polymerase inhibitors as radiosensitizers: a systematic review of pre-clinical and clinical human studies. Oncotarget 2017;8:69105-24. [Crossref] [PubMed]
- Ohmoto A, Yachida S. Current status of poly(ADP-ribose) polymerase inhibitors and future directions. Onco Targets Ther 2017;10:5195-208. [Crossref] [PubMed]
- Zhu A, Zhang CX, Lieberman HB. Rad9 has a functional role in human prostate carcinogenesis. Cancer Res 2008;68:1267-74. [Crossref] [PubMed]
- Cheng CK, Chow LW, Loo WT, et al. The cell cycle checkpoint gene Rad9 is a novel oncogene activated by 11q13 amplification and DNA methylation in breast cancer. Cancer Res 2005;65:8646-54. [Crossref] [PubMed]
- Broustas CG, Lieberman HB. RAD9 enhances radioresistance of human prostate cancer cells through regulation of ITGB1 protein levels. Prostate 2014;74:1359-70. [Crossref] [PubMed]
- Broustas CG, Zhu A, Lieberman HB. Rad9 protein contributes to prostate tumor progression by promoting cell migration and anoikis resistance. J Biol Chem 2012;287:41324-33. [Crossref] [PubMed]
- Frisch SM, Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. J Cell Biol 1994;124:619-26. [Crossref] [PubMed]
- Wang L, Hsu CL, Ni J, et al. Human checkpoint protein hRad9 functions as a negative coregulator to repress androgen receptor transactivation in prostate cancer cells. Mol Cell Biol 2004;24:2202-13. [Crossref] [PubMed]
- Hsu CL, Chen YL, Ting HJ, et al. Androgen receptor (AR) NH2- and COOH-terminal interactions result in the differential influences on the AR-mediated transactivation and cell growth. Mol Endocrinol 2005;19:350-61. [Crossref] [PubMed]
- Moehren U, Denayer S, Podvinec M, et al. Identification of androgen-selective androgen-response elements in the human aquaporin-5 and Rad9 genes. Biochem J 2008;411:679-86. [Crossref] [PubMed]
- Chlenski A, Nakashiro K, Ketels KV, et al. Androgen receptor expression in androgen-independent prostate cancer cell lines. Prostate 2001;47:66-75. [Crossref] [PubMed]
- Hang H, Lieberman HB. Physical interactions among human checkpoint control proteins HUS1p, RAD1p, and RAD9p, and implications for the regulation of cell cycle progression. Genomics 2000;65:24-33. [Crossref] [PubMed]
- Doré AS, Kilkenny ML, Rzechorzek NJ, et al. Crystal structure of the rad9-rad1-hus1 DNA damage checkpoint complex--implications for clamp loading and regulation. Mol Cell 2009;34:735-45. [Crossref] [PubMed]
- Sohn SY, Cho Y. Crystal structure of the human rad9-hus1-rad1 clamp. J Mol Biol 2009;390:490-502. [Crossref] [PubMed]
- Xu M, Bai L, Gong Y, et al. Structure and functional implications of the human rad9-hus1-rad1 cell cycle checkpoint complex. J Biol Chem 2009;284:20457-61. [Crossref] [PubMed]
- Yin Y, Zhu A, Jin YJ, et al. Human RAD9 checkpoint control/proapoptotic protein can activate transcription of p21. Proc Natl Acad Sci USA 2004;101:8864-9. [Crossref] [PubMed]
- Hopkins KM, Auerbach W, Wang XY, et al. Deletion of mouse rad9 causes abnormal cellular responses to DNA damage, genomic instability, and embryonic lethality. Mol Cell Biol 2004;24:7235-48. [Crossref] [PubMed]
- Zhu A, Zhou H, Leloup C, et al. Differential impact of mouse Rad9 deletion on ionizing radiation-induced bystander effects. Radiat Res 2005;164:655-61. [Crossref] [PubMed]
- Lieberman HB, Hopkins KM, Nass M, et al. A human homolog of the Schizosaccharomyces pombe rad9+ checkpoint control gene. Proc Natl Acad Sci U S A 1996;93:13890-5. [Crossref] [PubMed]
- Dang T, Bao S, Wang XF. Human Rad9 is required for the activation of S-phase checkpoint and the maintenance of chromosomal stability. Genes Cells 2005;10:287-95. [Crossref] [PubMed]
- Zou L, Liu D, Elledge SJ. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc Natl Acad Sci USA 2003;100:13827-32. [Crossref] [PubMed]
- Cotta-Ramusino C, McDonald ER 3rd, Hurov K, et al. A DNA damage response screen identifies RHINO, a 9-1-1 and TopBP1 interacting protein required for ATR signaling. Science 2011;332:1313-7. [Crossref] [PubMed]
- Lindsey-Boltz LA, Kemp MG, Capp C, et al. RHINO forms a stoichiometric complex with the 9-1-1 checkpoint clamp and mediates ATR-Chk1 signaling. Cell Cycle 2015;14:99-108. [Crossref] [PubMed]
- Sierant ML, Archer NE, Davey SK. The Rad9A checkpoint protein is required for nuclear localization of the claspin adaptor protein. Cell Cycle 2010;9:548-56. [Crossref] [PubMed]
- Liu S, Song N, Zou L. The conserved C terminus of Claspin interacts with Rad9 and promotes rapid activation of Chk1. Cell Cycle 2012;11:2711-6. [Crossref] [PubMed]
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 2008;9:616-27. [Crossref] [PubMed]
- Pandita RK, Sharma GG, Laszlo A, et al. Mammalian Rad9 plays a role in telomere stability, S- and G2-phase-specific cell survival, and homologous recombinational repair. Mol Cell Biol 2006;26:1850-64. [Crossref] [PubMed]
- Balakrishnan L, Brandt PD, Lindsey-Boltz LA, et al. Long patch base excision repair proceeds via coordinated stimulation of the multienzyme DNA repair complex. J Biol Chem 2009;284:15158-72. [Crossref] [PubMed]
- Gembka A, Toueille M, Smirnova E, et al. The checkpoint clamp, Rad9-Rad1-Hus1 complex, preferentially stimulates the activity of apurinic/apyrimidinic endonuclease 1 and DNA polymerase beta in long patch base excision repair. Nucleic Acids Res 2007;35:2596-608. [Crossref] [PubMed]
- Helt CE, Wang W, Keng PC, et al. Evidence that DNA damage detection machinery participates in DNA repair. Cell Cycle 2005;4:529-32. [Crossref] [PubMed]
- Hwang BJ, Jin J, Gunther R, et al. Association of the Rad9-Rad1-Hus1 checkpoint clamp with MYH DNA glycosylase and DNA. DNA Repair 2015;31:80-90. [Crossref] [PubMed]
- Panigrahi SK, Hopkins KM, Lieberman HB. Regulation of NEIL1 protein abundance by RAD9 is important for efficient base excision repair. Nucleic Acids Res 2015;43:4531-46. [Crossref] [PubMed]
- Li T, Wang Z, Zhao Y, et al. Checkpoint protein Rad9 plays an important role in nucleotide excision repair. DNA Repair 2013;12:284-92. [Crossref] [PubMed]
- He W, Zhao Y, Zhang C, et al. Rad9 plays an important role in DNA mismatch repair through physical interaction with MLH1. Nucleic Acids Res 2008;36:6406-17. [Crossref] [PubMed]
- Tsai FL, Kai M. The checkpoint clamp protein Rad9 facilitates DNA-end resection and prevents alternative non-homologous end joining. Cell Cycle 2014;13:3460-4. [Crossref] [PubMed]
- Crespan E, Czabany T, Maga G, et al. Microhomology-mediated DNA strand annealing and elongation by human DNA polymerases λ and β on normal and repetitive DNA sequences. Nucleic Acids Res 2012;40:5577-90. [Crossref] [PubMed]
- Komatsu K, Hopkins KM, Lieberman HB, et al. Schizosaccharomyces pombe Rad9 contains a BH3-like region and interacts with the anti-apoptotic protein Bcl-2. FEBS Lett 2000;481:122-6. [Crossref] [PubMed]
- Komatsu K, Miyashita T, Hang H, et al. Human homologue of S. pombe Rad9 interacts with BCL-2/BCL-xL and promotes apoptosis. Nat Cell Biol 2000;2:1-6. [Crossref] [PubMed]
- Yoshida K, Komatsu K, Wang HG, et al. c-Abl tyrosine kinase regulates the human Rad9 checkpoint protein in response to DNA damage. Mol Cell Biol 2002;22:3292-300. [Crossref] [PubMed]
- Yoshida K, Wang HG, Miki Y, et al. Protein kinase Cdelta is responsible for constitutive and DNA damage-induced phosphorylation of Rad9. EMBO J 2003;22:1431-41. [Crossref] [PubMed]
- Lieberman HB, Panigrahi SK, Hopkins KM, et al. p53 and RAD9, the DNA Damage Response, and Regulation of Transcription Networks. Radiat Res 2017;187:424-32. [Crossref] [PubMed]
- Ishikawa K, Ishii H, Murakumo Y, et al. Rad9 modulates the P21WAF1 pathway by direct association with p53. BMC Mol Biol 2007;8:37. [Crossref] [PubMed]
- el-Deiry WS, Kern SE, Pietenpol JA, et al. Definition of a consensus binding site for p53. Nat Genet 1992;1:45-9. [Crossref] [PubMed]
- Wang B, Xiao Z, Ren EC. Redefining the p53 response element. Proc Natl Acad Sci USA 2009;106:14373-8. [Crossref] [PubMed]
- Tebaldi T, Zaccara S, Alessandrini F, et al. Whole-genome cartography of p53 response elements ranked on transactivation potential. BMC Genomics 2015;16:464. [Crossref] [PubMed]
- Wen FC, Chang TW, Tseng YL, et al. hRAD9 functions as a tumor suppressor by inducing p21-dependent senescence and suppressing epithelial-mesenchymal transition through inhibition of Slug transcription. Carcinogenesis 2014;35:1481-90. [Crossref] [PubMed]
- Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol 2002;2:569-79. [Crossref] [PubMed]
- Lee MW, Hirai I, Wang HG. Caspase-3-mediated cleavage of Rad9 during apoptosis. Oncogene 2003;22:6340-6. [Crossref] [PubMed]
- Meyerkord CL, Takahashi Y, Araya R, et al. Loss of Hus1 sensitizes cells to etoposide-induced apoptosis by regulating BH3-only proteins. Oncogene 2008;27:7248-59. [Crossref] [PubMed]


