
cMET-N375S germline mutation is associated with poor prognosis of melanoma in Chinese patients
Introduction
Melanoma is a highly malignant tumor with rapid development and poor prognosis (1). In recent years, almost 200,000 new cases of melanoma were diagnosed globally, and an estimated 50,000 deaths occurred due to the disease (2). Despite the extensively study results from western countries, knowledge of melanoma in Asian patients is scarce. Different from the rates in Caucasians, acral and mucosal melanomas are the common types in most Asian patients with melanoma. In an analysis of 522 Asian patients diagnosed with malignant melanoma, these two subtypes are almost accounted for 65% of all patients (3). However, acral and mucosal melanomas showed a markedly different genomic landscape from cutaneous melanoma, with a far lower mutation burden dominated by large-scale structural variants (4). Thus, while the individualized targeted therapy has achieved success in recent years, a lot of Chinese patients with melanoma still do not have validated targets for targeted therapy.
cMET, a receptor tyrosine kinase (RTK) for hepatocyte growth factor (HGF), has been shown to play a key role in malignant melanoma tumor progression and is significantly related to patient survival (5-7). HGF is a mitogen of human melanocytes and contributes to the acquisition of the invasive phenotype in melanoma (5). In many cancer cells, cMET signaling can be constitutively activated through HGF-induced paracrine and autocrine signaling and through the presence of cMET mutations or protein overexpression (6,8-10). Several studies have shown that cMET can induce the activation of downstream signal transduction pathways, including the phosphatidylinositol 3-kinase/Akt and mitogen-activated protein kinase pathways, which are key targets for melanoma (11,12). Due to this activation mechanism, immediate resistance to BRAF (13-15) and NRAS (16) inhibitors can occur in melanoma cells. Thus, cMET is a potential new therapeutic target for melanoma treatment (17,18).
cMET consists of a heterodimer with 50-kDa α chain and 140-kDa β chain (19). As shown in Figure 1A, the canonical human cMET family includes the Sema (semaphorins), PSI (plexins, semaphorins, integrins), four IPT repeat (immunoglobulins, plexins, transcription factors), transmembrane (TM), juxtamembrane (JM) and tyrosine kinase (TK) domains (20). Among them, extracellular N-terminal 500 residues fold into a Sema domain, which is necessary for HGF ligand binding and receptor dimerization. A number of nonsynonymous mutations have been shown to be localized to this ligand binding domain in many cancers (10,21).
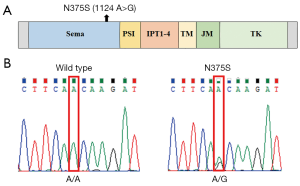
In a previous study of lung cancer, the N375S germline mutation was found to occur at a higher frequency in East Asians than Caucasians (13% vs. 1%) (21). It has been reported that this germline missense variation may decrease susceptibility to gastric cancer in a relatively large Chinese population (22). But the status of this mutation in melanoma is unknown. Therefore, in this study, we aim to examine the frequency of cMET-N375S and correlate it with phospho-cMET levels and other frequently mutated genes, such as BRAF and NRAS. We also evaluated correlations of cMET-N375S mutation with clinicopathological parameters and prognosis in patients with melanoma. This study may provide potential therapeutic targets for melanoma patients with cMET-N375S mutation.
Methods
Patients and tumor tissue samples
Paraffin-embedded tissue specimens from 181 patients with melanoma were obtained from Peking University Cancer Hospital & Institute. These samples were analyzed by hematoxylin and eosin (H&E) staining and by immunohistochemistry to confirm the diagnosis of melanoma. Clinical data, including age, sex, stage, thickness, ulceration, and survival (follow-up through October 2015), were collected. For patients during stage II/III, surgical resection and high-dose interferon adjuvant treatment are the main therapeutic regimens. For stage IV patients, the main types of therapies are dacarbazine-based chemotherapy regimens. No more than 5% of patients were treated with BRAF inhibitors or immunotherapies. The study was approved by the Medical Ethics Committee of the Peking University Cancer Hospital & Institute.
DNA extraction and mutation analysis
We extracted genomic DNA from formalin-fixed, paraffin-embedded (FFPE) sections using a QIAamp DNA FFPE Tissue Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. We amplified exon 2 of the cMET gene, exons 11 and 15 of the BRAF gene, exons 1 and 2 of the NRAS gene and exons 9, 11, 13, 15 and 17 of the CKIT gene by polymerase chain reaction (PCR) in three separate preparations of genomic DNA. The primer sequences are listed in Table S1. We purified PCR products with QIAquick (Qiagen), and directly sequenced them using Big Dye Terminator sequencing chemistry on an ABI3130 automated sequencer (Applied Biosystems, Foster City, CA, USA). All aberrations were verified by repeated bidirectional sequencing on the ABI sequencer. In the case of single nucleotide polymorphisms, blood samples were obtained, and genomic DNA was extracted from peripheral lymphocytes using standard methods.
Immunohistochemistry
Immunohistochemical (IHC) analyses were performed using antibodies against p-Met (Cell Signaling Technology, Beverly, MA, USA), followed by a standard avidin-biotin detection protocol using 3-amino-9 ethylcarbazole (AEC). The staining score for each sample, based on the intensity and density of the staining, was graded as 0, 1, 2, and 3 (0 as negative and 3 as strongest; or 0 as negative and 1, 2, and 3 as positive, respectively) by three pathologists independently, without knowledge of the cMET mutation status of these patients.
Statistical methods for clinical correlation
SPSS 16.0 software was used for all statistical analyses. Categorical data, such as sex, were described using frequencies and percentages. Continuous data, such as age and thickness, were described using means ± standard deviations for normally distributed data. We used unpaired t-tests to evaluate differences in measurement data of two groups and Pearson χ2 tests, Fisher’s exact tests, or Kruskal-Wallis tests to analyze the correlation between clinicopathological parameters and different groups. Overall survival (OS) and disease-free survival (DFS) curves were calculated according to the Kaplan-Meier method, and comparisons were performed using log-rank tests. All statistical analyses were two sided, and differences with P values of less than 0.05 were considered statistically significant.
Results
cMET mutations in melanoma subtypes
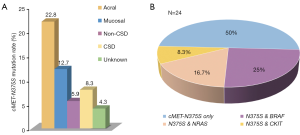
Of the 181 melanoma samples analyzed, 26 (14.4%) were found to contain nonsynonymous cMET mutations. With only two exceptions (C788T and C463A mutations), all mutations were N375S [24 (13.26%), Figure 1B]. The frequencies of cMET-N375S mutations in acral and mucosal melanoma, melanomas of the skin without chronic sun induced damage (non-CSD), melanomas of the skin with chronic sun induced damage (CSD) melanoma, and unknown primary subtypes were 22.8%, 12.7%, 5.9%, 8.3%, and 4.3%, respectively (Figure 2A). The N375S mutation was found at a higher frequency in acral and mucosal melanomas (20/112, 17.9%) than in other types of melanomas (4/69, 5.8%; P=0.036). Consistent with previous conclusions (21), this N375S mutation could be found in peripheral blood lymphocyte DNA, indicating that it was a germline polymorphism.
Correlation of the cMET-N375S mutation with clinicopathological parameters of melanoma
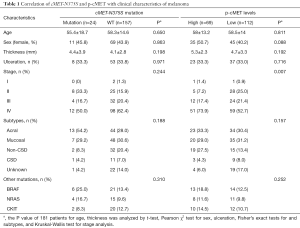
In our cohort, we analyzed the correlations of cMET-N375S mutations with the characteristics of patients with melanoma, including age, sex, ulceration, thickness, and clinical stage (Table 1). There were no significant differences between patients with melanoma with or without cMET-N375S mutation in terms of age, sex, and clinical stage.
Full table
Ulceration and the Breslow thickness of melanomas are important prognostic indicators. Since most patients’ data regarding mitotic rate, vascular invasion and degree of tumor-infiltrating lymphocytes were not available, we did not analyze these clinical features. Among the 181 samples with data available, the average thickness of samples with cMET-N375S mutations was 4.4±3.9 mm, whereas that of samples without this mutation was 4.1±2.8 mm (P=0.198). Additionally, the ulceration rate was not significantly different between patients with and without cMET-N375S mutations (P=0.971).
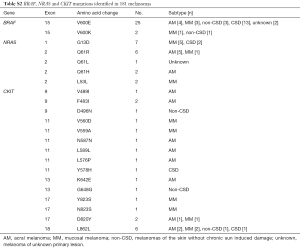
Next, to elucidate the relationships between the type and frequency of cMET mutations with other common mutations in melanoma, we also examined mutations in well-established target genes (e.g., BRAF, CKIT, and NRAS) in the 181 melanoma samples (Table 1). BRAF, NRAS and CKIT mutations were found in 27 (14.9%), 19 (10.5%), 22 (12.1%) of our cohort, respectively (Table S2). Among the 24 cases with cMET-N375S mutations, six cases were found to harbor the BRAF-V600E mutation, four cases harbored NRAS mutations (one case of G12C, one case of L53L, one case of Q61R, and one case of Q61H), and two cases harbored CKIT mutations (one case of L589L, one case of L862L) (Figure 2B). Further analysis of the frequencies of BRAF and NRAS mutations in patients of our cohort showed that the mutation frequencies of BRAF and NRAS did differ significantly between patients with or without cMET-N375S mutations (P=0.039).
Effects of cMET-N375S mutation on the prognosis of melanoma
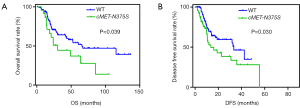
We then analyzed the prognostic significance of the cMET-N375S mutation for OS (Figure 3A). The OS rates were determined for 181 patients. The results showed that the median follow-up period was 24 (range, 4–231) months, and the median survival time for patients with cMET-N375S [30.5 months; 95% confidence interval (CI), 6.6–105.2 months] was significantly shorter than that for patients without the nonsynonymous cMET-N375S mutation (65.9 months; 95% CI, 3.9–136.9 months; P=0.039). Therefore, the cMET-N375S mutation may be an important prognostic factor for patients with melanoma, and melanoma patients with this mutation may show higher risk of death. In multivariate Cox regression assays (Table 2), the N375S mutation was validated as an independent prognostic factor for OS [P=0.003, hazard ratio (HR) =3.557]. Also, cMET-N375S was associated with DFS (P=0.030, Figure 3B). The DFS rates were determined for 176 patients. The median survival time for patients with cMET-N375S (13.6 months) was significantly shorter than that for patients without the nonsynonymous cMET-N375S mutation (33.3 months).

Full table
p-cMET levels in melanoma subtypes
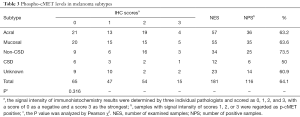
p-cMET levels were divided into high (IHC score =2 or 3) and low (IHC score =0 or 1) groups (Figure 4). Patient and clinic characteristics based on p-cMET levels are summarized in Table 1. There were statistically significant differences in stage (P=0.007) between patients with high or low levels of p-cMET.
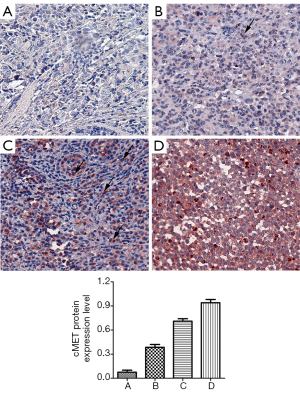
Among the 181 samples, the overall rate of detection of p-cMET staining was 64.1% (116/181). Positive p-cMET staining was observed in 63.2% (36/57) of acral, 63.6% (35/55) of mucosal, 73.5% (25/34) of non-CSD, 50.0% (6/12) of CSD, and 60.9% (14/23) of unknown primary site melanomas. We found that p-cMET levels were not significantly different between these subtypes (P=0.316; Table 3).
Full table
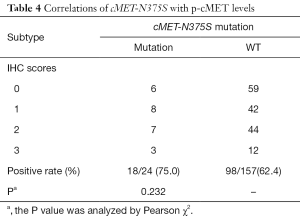
Next, we analyzed the correlation between cMET-N375S mutations and cMET expression levels. Among the 24 cases with cMET-N375S mutations, the IHC detection rate for p-cMET was 75.0% (18/24), which was not significantly higher (P=0.232) than that in cases without N375S mutation (62.4%, 98/157; Table 4).
Full table
Discussion
In a previous study of lung cancer, the N375S germline mutation was found to occur at a higher frequency in East Asians than Caucasians (13% vs. 1%) (21). Here, we found that the frequency of the cMET-N375S germline mutation was 13.26% (24/181) in patients with melanoma in our cohort.
Different from Caucasians, acral and mucosal melanomas are the common types of melanoma diagnosed in Asian patients. The mechanism, clinical features and treatment principle of mucosal and acral melanomas are different from those of cutaneous melanoma. The principal mutation mechanisms driving these two subtypes of melanoma were not attributable to ultraviolet radiation and imply novel carcinogenic exposures (4). Moreover, we found that the incidence of the cMET-N375S mutation was much higher in patients with acral and mucosal melanomas than in patients with other subtypes. According to our findings, it is suggested that larger sample size and further research is necessary to clarify potential links between cMET-N375S mutation and these two special subtypes of melanoma in China.
Resistance to vemurafenib (a first-in-class small-molecule BRAF inhibitor) has become a major issue in patients with melanoma. cMET activation in melanoma cell lines shows primary resistance to vemurafenib, and BRAF/cMET combination therapy results in reversal of vemurafenib resistance (13,14). In addition, the Sema domain of cMET harbors the ligand binding site; the effects of mutations in this domain on the function of the protein should be assessed. And, replacement of asparagine 375 with serine can disrupt HGF ligand binding by altering the molecular structure. Thus, cMET-N375S mutation leads to the emergence of Met inhibitor resistance (23). Furthermore, we found that the cMET-N375S mutation often co-existed with BRAF mutations. Accordingly, we suggest that alterations in BRAF and elevated p-cMet may be strong indications for combination therapy, although careful attention is required to exclude cMET-N375S mutation.
Next, we intended to find out the relationship between this mutation and the prognosis of Chinese melanoma patients. cMET-N375S was found to decrease gastric cancer susceptibility (22) and was not shown to be associated with lung cancer prognosis (23). However, our data showed that OS and DFS differed significantly between patients with and without the cMET-N375S mutation. This may due to differences in cancer type. Thus, additional large-scale studies are needed to clarify the effects of the cMET-N375S mutation on cancer prognosis, especially in acral and mucosal melanoma.
We also evaluated the p-cMET expression levels in 181 patients. Several studies have shown correlation between cMET expression and progression or aggressive behavior of many cancers (9,10,24). Moreover, a previous study showed that cMET phosphorylation was twice as high in patients with melanoma than in patients with nevus. Though we find out that cMET-N375S mutation is associated with poor prognosis of Chinese melanoma patients, there was no correlation between this mutation and p-cMET levels (P=0.232). The accurate molecular mechanisms remain unclear.
Conclusions
Our study demonstrated that the cMET-N375S germline mutation was an independent adverse prognostic factor for melanoma. Although we did not perform functional experiments in vivo or in vitro, our study provided clinical data to support further investigations on cMET as a potential therapeutic target in Chinese patients with melanoma.

Full table
Full table
Acknowledgments
Funding: This work was supported by grants from National Natural Science Foundation of China (81672696, 81772912), Beijing Municipal Natural Science Foundation (7152033), Beijing Baiqianwan Talents Project, Beijing Municipal Administration of Hospitals Clinical medicine Development of special funding support (ZYLX201603), Beijing Talents Fund (2016000021223ZK18) and Beijing Municipal Science & Technology Commission (Z151100003915074).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.02.13). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study protocol was approved by the Medical Ethics Committee of the Peking University Cancer Hospital & Institute. All procedures performed in studies involving human participants were in accordance with the ethical standards of Peking University Cancer Hospital & Institute and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Written informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature 2007;445:851-7. [Crossref] [PubMed]
- Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010;127:2893-917. [Crossref] [PubMed]
- Chi Z, Li S, Sheng X, et al. Clinical presentation, histology, and prognoses of malignant melanoma in ethnic Chinese: a study of 522 consecutive cases. BMC Cancer 2011;11:85. [Crossref] [PubMed]
- Hayward NK, Wilmott JS, Waddell N, et al. Whole-genome landscapes of major melanoma subtypes. Nature 2017;545:175-80. [Crossref] [PubMed]
- Natali PG, Nicotra MR, Di Renzo MF, et al. Expression of the c-Met/HGF receptor in human melanocytic neoplasms: demonstration of the relationship to malignant melanoma tumour progression. Br J Cancer 1993;68:746-50. [Crossref] [PubMed]
- Park M, Dean M, Cooper CS, et al. Mechanism of met oncogene activation. Cell 1986;45:895-904. [Crossref] [PubMed]
- Halaban R, Rubin JS, Funasaka Y, et al. Met and hepatocyte growth factor/scatter factor signal transduction in normal melanocytes and melanoma cells. Oncogene 1992;7:2195-206. [PubMed]
- Jeffers M, Schmidt L, Nakaigawa N, et al. Activating mutations for the met tyrosine kinase receptor in human cancer. Proc Natl Acad Sci U S A 1997;94:11445-50. [Crossref] [PubMed]
- Seiwert TY, Jagadeeswaran R, Faoro L, et al. The MET receptor tyrosine kinase is a potential novel therapeutic target for head and neck squamous cell carcinoma. Cancer Res 2009;69:3021-31. [Crossref] [PubMed]
- Ma PC, Jagadeeswaran R, Jagadeesh S, et al. Functional expression and mutations of c-Met and its therapeutic inhibition with SU11274 and small interfering RNA in non-small cell lung cancer. Cancer Res 2005;65:1479-88. [Crossref] [PubMed]
- Furge KA, Zhang YW, Vande Woude GF. Met receptor tyrosine kinase: enhanced signaling through adapter proteins. Oncogene 2000;19:5582-9. [Crossref] [PubMed]
- Ma PC, Tretiakova MS, Nallasura V, et al. Downstream signalling and specific inhibition of c-MET/HGF pathway in small cell lung cancer: implications for tumour invasion. Br J Cancer 2007;97:368-77. [Crossref] [PubMed]
- Vergani E, Vallacchi V, Frigerio S, et al. Identification of MET and SRC activation in melanoma cell lines showing primary resistance to PLX4032. Neoplasia 2011;13:1132-42. [Crossref] [PubMed]
- Straussman R, Morikawa T, Shee K, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature 2012;487:500-4. [Crossref] [PubMed]
- Byeon HK, Na HJ, Yang YJ, et al. c-Met-mediated reactivation of PI3K/AKT signaling contributes to insensitivity of BRAF(V600E) mutant thyroid cancer to BRAF inhibition. Mol Carcinog 2016;55:1678-87. [Crossref] [PubMed]
- Chattopadhyay C, Ellerhorst JA, Ekmekcioglu S, et al. Association of activated c-Met with NRAS-mutated human melanomas. Int J Cancer 2012;131:E56-65. [Crossref] [PubMed]
- Puri N, Ahmed S, Janamanchi V, et al. c-Met is a potentially new therapeutic target for treatment of human melanoma. Clin Cancer Res 2007;13:2246-53. [Crossref] [PubMed]
- Abdel-Rahman MH, Boru G, Massengill J, et al. MET oncogene inhibition as a potential target of therapy for uveal melanomas. Invest Ophthalmol Vis Sci 2010;51:3333-9. [Crossref] [PubMed]
- Faletto DL, Tsarfaty I, Kmiecik TE, et al. Evidence for non-covalent clusters of the c-met proto-oncogene product. Oncogene 1992;7:1149-57. [PubMed]
- Ma PC, Maulik G, Christensen J, et al. c-Met: structure, functions and potential for therapeutic inhibition. Cancer Metastasis Rev 2003;22:309-25. [Crossref] [PubMed]
- Krishnaswamy S, Kanteti R, Duke-Cohan JS, et al. Ethnic differences and functional analysis of MET mutations in lung cancer. Clin Cancer Res 2009;15:5714-23. [Crossref] [PubMed]
- Liu Y, Zhang Q, Ren C, et al. A germline variant N375S in MET and gastric cancer susceptibility in a Chinese population. J Biomed Res 2012;26:315-8. [Crossref] [PubMed]
- Shieh JM, Tang YA, Yang TH, et al. Lack of association of C-Met-N375S sequence variant with lung cancer susceptibility and prognosis. Int J Med Sci 2013;10:988-94. [Crossref] [PubMed]
- Jagadeeswaran R, Ma PC, Seiwert TY, et al. Functional analysis of c-Met/hepatocyte growth factor pathway in malignant pleural mesothelioma. Cancer Res 2006;66:352-61. [Crossref] [PubMed]


