
PD-1 in cancer: Dr. Jekyll and Mr. Hyde
T cell non-Hodgkin lymphomas (NHL) represent about 6% of non-cutaneous lymphomas and 65% of cutaneous lymphomas (1). Recent insights into the genetic landscape of nodal T cell NHL have led to an update in their classification (2). Despite this advance, peripheral T cell lymphomas (PTCL) still show morphologic, phenotypic, genetic and prognostic heterogeneity. Combinations of deep sequencing and transcriptomic analyses have identified mutations of epigenetic regulators, tumor suppressor genes and, interestingly, gain of function mutations of the T cell receptor (TCR) pathway (3,4).
Despite intensive chemotherapy regimens, the outcome is poor, with a 5-year overall survival around 30% (5). Thus, PTCL depict a large unmet medical need and various clinical trials are going on. Great hopes are based on immune checkpoint inhibitors whose leaders anti PD-1 and anti-CTL4 antibodies have shown clinical efficacy in solid tumors and Hodgkin lymphomas through the activation of T cells antitumor immunity (6). In a recent Nature paper, however, Wartewig and colleagues demonstrate that PD-1 plays a surprising role as haploinsufficient tumor suppressor in T cell lymphomas, thus implicating that anti PD-1 antibodies would rather be a detrimental therapy in T cell NHL (7).
In this work, the authors investigated a murine model whereby T cell NHL is driven by the recurrent chromosomal translocation t(5-9) (q33;q22). This translocation is described in up to 17% of human PTCL (8). The resulting IL-2 inducible T cell kinase (ITK)—spleen tyrosine kinase (SYK) fusion protein is a constitutively active kinase that mimics the TCR signal and drives lymphomagenesis (9). Conditional expression of ITK-SYK in CD4+ T cells (ITK-SYKCD4-Cre mice) triggers the development of lymphomas comparable to human PTCL (9). Surprisingly, when analyzing the tamoxifen-inducible expression of ITK-SYK in CD4+ T cells (ITK-SYKCD4-CreERT2 mice), the authors observed that, after an initial proliferation phase following tamoxifen exposition, ITK-SYK expressing T cells significantly decreased. However, two thirds of the mice that received the highest dose of tamoxifen finally developed lymphomas after a longer follow-up of almost one year. This observation suggests that T cells may detain tumor suppressive properties that can be counteracted within time by acquisition of secondary oncogenic hits.
To address this hypothesis, the authors performed genome-wide mutagenesis using the PiggyBac transposition tool (10). Briefly, the PiggyBac transposons interrupt gene expression by inserting in or around genes in a random manner. Then semiquantitative transposon insertion site sequencing allows for identification of putative cancer driver genes (11). Following in vivo transposition in ITK-SYK CD4+ T cells, lymphomagenesis was accelerated and was associated to frequent insertions in the Pdcd1 locus, which codes for the inhibitory programmed death receptor 1 (PD-1). Consistently, lymphomas with Pdcd1 transposons insertions showed decreased PD-1 expression.
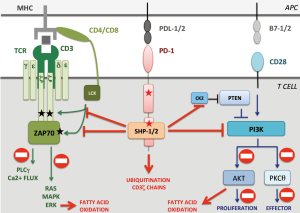
PD-1 is a transmembrane receptor that inhibits TCR signal transduction after binding with programmed death ligand 1 or 2 (PD-L1/2), which are members of the B7 family (12). The mechanisms by which engaged PD-1 leads to TCR signal down-modulation are partially understood. Briefly, engaged PD-1 is enrolled in the immunological synapse and recruits SHP1 and SHP2 phosphatases to counteract ZAP70 and PI3K activation. Furthermore, engaged PD-1 inhibits CK2 resulting in PTEN activation, which further blocks PI3K-AKT-PKCθ pathway and decreases T cell proliferation and survival (13) (Figure 1).
Wartewig et al. analyzed 158 cases of human T cell lymphomas, mostly cutaneous lymphomas, and found 23% of Pdcd1 alterations. They were almost exclusively homo or heterozygous deletions associated with decreased Pdcd1 mRNA expression, making PD-1 a good tumor suppressor candidate in T cell lymphomagenesis.
To further explore this hypothesis, the authors next demonstrated that PD-1 expression is upregulated by oncogenic activation of the TCR pathway and could act as a negative feedback loop of lymphomagenesis. Indeed, when crossing ITK-SYKCD4-CreERT2 with PD-1−/− mice, ITK-SYK expressing T cells underwent massive and uncontrolled proliferation after tamoxifen injection, leading to animals’ death within only 1 week. This accelerated lymphoproliferative disease was transmitted to recipient animals after transplantation of ITK-SYK PD-1− CD4+ T cells. Fatal transplantable lymphomas were observed also when ITK-SYKCD4-CreERT2 mice were crossed with PD-1+/− animals, mimicking Pdcd1 heterozygous deletions observed in human T cell lymphomas. This demonstrates that Pdcd1 acts as a haploinsufficient tumor suppressor gene in T cell NHL. Interestingly, PD-1 deletions alone are not sufficient to induce T cell lymphomagenesis as PD-1−/− knockout mice are known to develop characteristic lupus-like autoimmune disease but do not experience lymphoma emergence (14), contrary to CTLA-4−/− knockout mice who die from lymphoproliferative disorder (15). This emphasizes that PD-1 knockout’s role in lymphomagenesis depends on the associated oncogenic TCR activation.
From a mechanistic point of view, the authors suggest that PD-1 deletions could contribute to T cell lymphomagenesis by inactivating PTEN, thus raising PI3K-AKT-PKCθ signaling. However, one could ask why these tumors delete PD-1 instead of deleting PTEN or selecting gain of function mutations of the PI3K pathway. A possible explanation is that PD-1 loss is instrumental to sustain lymphomagenesis through unknown PI3K-independent mechanisms par example by directly inhibiting the TCR signaling or by promoting metabolic changes (Figure 1). Further investigations will thus be necessary to fully dissect the molecular mechanisms whereby PD-1 prevents T-cell lymphomagenesis.
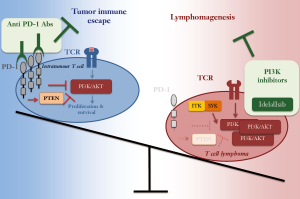
Nevertheless, the study by Wartewig and colleagues has important therapeutic implications. First, when treated with anti PD-1 or anti PD-L1 antibodies, ITK-SYKCD4-CreERT2 mice quickly developed aggressive T cell lymphomas. Nevertheless, after transplantation in non-antibody treated recipients, these lymphomas failed to grow. Therefore, checkpoint inhibitors could transiently enhance the proliferation of T cells with oncogenic activation of the TCR pathway. As such, in some T cell lymphomas PD-1 antibodies not only would not represent a therapeutic option but they would even exert detrimental consequences.
Second, the PI3K inhibitor idelalisib, which is approved for the treatment of chronic lymphocytic leukemia and follicular lymphoma, significantly improved the survival of mice transplanted with tamoxifen-induced ITK-SYKCD4-CreERT2; PD-1−/− T cells. Albeit lacking independent confirmations with a genetic approach (16), these pre-clinical data are intriguing as they suggest that PI3K inhibitors would be valuable treatment in T cell lymphomas.
To conclude, by demonstrating an unknown tumor suppressor role for PD-1, Wartewig’s work reveals that PD-1 role in cancer development is dual and strictly depending on the expression levels and the cellular context (Figure 2). In infiltrating tumor cells, elevated PD-1 levels are necessary to suppress T cell proliferation and survival, thereby promoting tumor immune escape. This context thus calls for anti PD-1 antibodies as novel therapies to boost the immune response against the tumor. In T cell lymphomas, instead, low PD-1 levels are instrumental to sustain lymphomagenesis by supporting the oncogenic TCR signaling. From the translational point of view, this suggests that a careful evaluation of tumor microenvironment, oncogenic TCR alterations and Pdcd1 genomic deletions are essential pre-requisites for evaluating the feasibility of anti PD-1 therapies in T cell NHL.
Acknowledgments
Funding: MT was supported by a grant from the “Fondation de France” and the “Fondation ARC”.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Chen Qian (Center for Inflammation & Epigenetics, Houston Methodist Hospital Research Institute, Houston, TX, USA).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.03.08). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Laurent C, Baron M, Amara N, et al. Impact of Expert Pathologic Review of Lymphoma Diagnosis: Study of Patients From the French Lymphopath Network. J Clin Oncol 2017;35:2008-17. [Crossref] [PubMed]
- Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016;127:2375-90. [Crossref] [PubMed]
- Palomero T, Couronné L, Khiabanian H, et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat Genet 2014;46:166-70. [Crossref] [PubMed]
- Vallois D, Dobay MP, Morin RD, et al. Activating mutations in genes related to TCR signaling in angioimmunoblastic and other follicular helper T-cell-derived lymphomas. Blood 2016;128:1490-502. [Crossref] [PubMed]
- Weisenburger DD, Savage KJ, Harris NL, et al. Peripheral T-cell lymphoma, not otherwise specified: a report of 340 cases from the International Peripheral T-cell Lymphoma Project. Blood 2011;117:3402-8. [Crossref] [PubMed]
- Batlevi CL, Matsuki E, Brentjens RJ, et al. Novel immunotherapies in lymphoid malignancies. Nat Rev Clin Oncol 2016;13:25-40. [Crossref] [PubMed]
- Wartewig T, Kurgyis Z, Keppler S, et al. PD-1 is a haploinsufficient suppressor of T cell lymphomagenesis. Nature 2017;552:121-5. [PubMed]
- Streubel B, Vinatzer U, Willheim M, et al. Novel t(5;9)(q33;q22) fuses ITK to SYK in unspecified peripheral T-cell lymphoma. Leukemia 2006;20:313-8. [Crossref] [PubMed]
- Pechloff K, Holch J, Ferch U, et al. The fusion kinase ITK-SYK mimics a T cell receptor signal and drives oncogenesis in conditional mouse models of peripheral T cell lymphoma. J Exp Med 2010;207:1031-44. [Crossref] [PubMed]
- Rad R, Rad L, Wang W, et al. PiggyBac transposon mutagenesis: a tool for cancer gene discovery in mice. Science 2010;330:1104-7. [Crossref] [PubMed]
- Friedrich MJ, Rad L, Bronner IF, et al. Genome-wide transposon screening and quantitative insertion site sequencing for cancer gene discovery in mice. Nat Protoc 2017;12:289-309. [Crossref] [PubMed]
- Riley JL. PD-1 signaling in primary T cells. Immunol Rev 2009;229:114-25. [Crossref] [PubMed]
- Arasanz H, Gato-Cañas M, Zuazo M, et al. PD1 signal transduction pathways in T cells. Oncotarget 2017;8:51936-45. [Crossref] [PubMed]
- Nishimura H, Nose M, Hiai H, et al. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity 1999;11:141-51. [Crossref] [PubMed]
- Waterhouse P, Penninger JM, Timms E, et al. Lymphoproliferative disorders with early lethality in mice deficient in Ctla-4. Science 1995;270:985-8. [Crossref] [PubMed]
- Swat W, Montgrain V, Doggett TA, et al. Essential role of PI3Kdelta and PI3Kgamma in thymocyte survival. Blood 2006;107:2415-22. [Crossref] [PubMed]


