
Editorial: fate mapping of human glioblastoma reveals an invariant stem cell hierarchy
Glioblastoma (GB) is the most aggressive primary brain cancer of adulthood, associated with an average 5-year survival rate of 2% (1). A small population of tumor-initiating cells, termed cancer stem cells (CSCs), are the putative origin of GB, accounting for tumor development, progression and recurrence (2).
There is a body of literature highlighting intra-tumoral heterogeneity of the CSC population within GB (3-7), suggesting the presence of multiple CSC phenotypes within the tumor. Whether these CSCs are derived from a single clone endowed with innate plasticity, inferring a CSC hierarchy (7), or these are independent clones (6), remains to be conclusively determined.
We have recently presented a review inferring the presence of a putative hierarchy of CSCs in GB (4), underscored by the CSC concept and the discovery of induced pluripotency by Takahashi et al. (8). We have also characterized CSCs within GB by their expression of the embryonic stem cell (ESC) markers NANOG, SOX2, SALL4, pSTAT3 and OCT4 at both the transcriptional and translational levels (3). Variations in the relative abundance of each marker were observed, with abundant and almost ubiquitous presence of the SOX2 protein and relatively low abundance of OCT4 expressed by fewer cells, consistent with their relative abundance at the transcriptional level (3).
We infer that the observed phenotypic heterogeneity within the CSC population within GB, results from a CSC hierarchy, with the least abundant OCT4+ subpopulation representing the most primitive cells at the apex, which differentiate to form the downstream more abundant SOX2+ progenitor cancer cells with a more restricted fate (4).
DNA barcoding methodologies have been utilized for both tracking cells and as a possible quantitative method of characterizing the population dynamics of cancer cells (9). By tracking barcoded cells derived from non-small cell lung cancer and chronic myeloid leukemia, Bhang et al. (9) have identified a small population of treatment-resistant clones, inferring the presence of a CSC population. Resistant clones account for 0.05% of the original cohort of barcoded non-small cell lung cancer cells, and 0.001% of barcoded cells derived from chronic myeloid leukemia. These observed growth patterns cannot be replicated through mathematical models anticipating high rates of genomic instability, thus suggesting the presence of a small subpopulation of pre-existing, highly tumorigenic cells, with innate treatment resistance (9). Lan et al. (10) propose a basic model of GB growth, by xenotransplantation of GB cells transduced with biologically neutral DNA barcodes into NOD/SCID/IL2Rˠ- mice. The resultant tumors histologically resemble GB and display both intra- and inter-tumoral heterogeneity, variations in proliferative activity, rates of apoptosis, growth and response to the chemotherapeutic agent temozolomide (TMZ). When cells derived from the tumor obtained from primary passage undergo xenotransplantation passaging, the number of unique surviving clones arising from approximately 3% of the barcoded cohort, diminish with each serial passage (10). This provides supporting evidence that tumorigenesis is perpetuated by a relatively small population of GB CSCs.
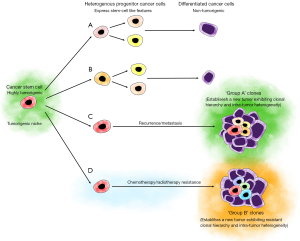
CSCs are the putative source of metastasis. Lan et al. (10) report correlations in barcoded clone sizes between ipsilateral and contralateral brain hemispheres. Xenografts derived from injecting clones dissociated from the contralateral hemisphere, generate clones which are mutual to both hemispheres at the earlier passage. In addition, across all passages and hemispheres, there is a consistent broad distribution of clone sizes which persistently exhibit a negative binomial distribution, even following treatment by TMZ (10). This growth behavior, functionally translates to the presence of a slow-cycling, self-renewing, primitive stem cell-like population, which presumably undergoes asymmetric cell division to give rise to rapidly proliferating progenitor cancer cells which ultimately differentiate to form non-renewing, short-lived differentiated cancer cells. These results parallel findings of Patel et al. (5) who establish a GB CSC signature based on the transcriptional profile of GB CSCs and differentiated GB cancer cells. This study shows that the stem-cell phenotype is not discrete and it lies on a continuum which is inversely correlated with cell-cycle transcriptional features, and positively correlated with expression of transcription factors concerning tumor propagation.
Treatment of xenografts with TMZ reveals two distinct behavioral phenotypes: a ‘Group A’ chemo-sensitive phenotype consisting the majority of the clones, and a ‘Group B’ chemo-resistant phenotype comprising the minority of clones. No correlation in clone sizes has been observed amongst Group A clones throughout serial passaging, while group B clone size positively correlates with passaging. The consistent negative binomial pattern of distribution of Group A clone sizes implies the chemo-resistant nature of CSC growth patterns. The emergence of Group B clones, suggests that a minority of the CSCs diverge from normal growth dynamics, and thus may be a prominent driver of the evolution of the CSC clonal hierarchy, and aberrant GB growth (10).
Lan et al. (10) demonstrate the putative presence of intra-tumoral CSC heterogeneity in GB, with only a small proportion being true CSCs possessing both the innate ability and plasticity to differentiate into a heterogeneous population of downstream progenitor cancer cells (Figure 1), some of which still display varying degrees of stemness. It is exciting to speculate that the phenotype of the dominant progenitor cancer cell population, prior to or following treatment, is dependent on the progenitors best suited to survive. Equally as intriguing, and which remains to be determined, is whether the more stem-like progenitor cells best suited to the environment, are able to upregulate ESC-associated genes, to become CSCs.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Xian-Xin Qiu (Shanghai Proton and Heavy Ion Center (SPHIC), a.k.a. the Proton and Heavy Ion Center of Fudan University Shanghai Cancer Center (FUSCC), Shanghai, China).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.03.19). T Itinteang and ST Tan are inventors of the PCT patent application (PCT/NZ2015/050108) Cancer Diagnosis and Therapy, and Cancer Therapeutic (PCT/NZ2018/050006). The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Surawicz TS, Davis F, Freels S, et al. Brain tumor survival: results from the National Cancer Data Base. J Neurooncol 1998;40:151-60. [Crossref] [PubMed]
- Vlashi E, Pajonk F. Cancer stem cells, cancer cell plasticity and radiation therapy. Semin Cancer Biol 2015;31:28-35. [Crossref] [PubMed]
- Bradshaw A, Wickremesekera A, Brasch HD, et al. Cancer Stem Cells in Glioblastoma Multiforme. Front Surg 2016;3:48. [PubMed]
- Bradshaw A, Wickremsekera A, Tan ST, et al. Cancer Stem Cell Hierarchy in Glioblastoma Multiforme. Front Surg 2016;3:21. [PubMed]
- Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014;344:1396-401. [Crossref] [PubMed]
- Sottoriva A, Spiteri I, Piccirillo SG, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A 2013;110:4009-14. [Crossref] [PubMed]
- Driessens G, Beck B, Caauwe A, et al. Defining the mode of tumour growth by clonal analysis. Nature 2012;488:527-30. [Crossref] [PubMed]
- Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007;131:861-72. [Crossref] [PubMed]
- Bhang HE, Ruddy DA, Krishnamurthy Radhakrishna V, et al. Studying clonal dynamics in response to cancer therapy using high-complexity barcoding. Nat Med 2015;21:440-8. [Crossref] [PubMed]
- Lan X, Jorg DJ, Cavalli FM, et al. Fate mapping of human glioblastoma reveals an invariant stem cell hierarchy. Nature 2017;549:227-32. [Crossref] [PubMed]


