
The overexpression of cytochrome c oxidase subunit 6C activated by Kras mutation is related to energy metabolism in pancreatic cancer
Introduction
RAS proteins are molecular switches that control cell growth and proliferation. Ras mutations are found in approximately 20–30% of all human cancers including most commonly three types of mutations, Hras, Nras and Kras. Kras is the most frequently isoform detected mutation occurring in about 90% of pancreatic cancers, about 40% colon rectal cancer and lung cancer (1,2). Kras mutations are characterized by single base missense mutations, 98% of which are found at residues G12, G13 or Q61 (the predominant substitution is G12D) (3). Ras mutations activate downstream signaling can promote tumorigenesis. Moreover, Kras mutation cancers are resistant to current therapies and often accompanied by poor prognosis and low survival rate. Kras mutations not only promote aerobic glycolysis and glutamine metabolism reprograming to provide energy (4,5), but also facilitate other branched metabolism pathways including autophagy and macropinocytosis (6,7). Since no directly targeted therapy currently exists for cancers with Kras mutations (8,9), targeting the enzymes involved in metabolic pathways may provide an alternative therapeutic approach for Kras mutation cancer.
In this study, we constructed a BxPC-3/KrasG12D cell line by using CRISPR/Cas9 system. By searching differential genes correlated with bioenergy metabolite in a pair of cells between BxPC-3/KrasG12D and wild type BxPC-3, we identified an up-regulated gene COX6C which encodes a subunits of the protein involved in respiration chain in mitochondrial inner membrane. We further characterized the function of COX6C in BxPC-3/KrasG12D cells in the context of mitochondrial function and cell viability. Our finding indicates COX6C might be as a potential new protein highly driven by KrasG12D mutant in pancreatic cancer.
Methods
Cell culture
The human pancreatic cancer BxPC-3 cell line (provided by the Cell Resource Center, Institute of Basic Medical Sciences, China) was cultured in RPMI-1640 medium (Hyclone; Thermo Fisher Scientific, Hudson, NH, USA) containing 10% heat-inactivated fetal bovine serum (FBS, Gibco; Life Technologies, Carlsbad, CA, USA) and antibiotics (100 U/mL penicillin and 100 µg/mL streptomycin) under humidified conditions with 5% (v/v) CO2 at 37 °C.
Establishment of stable KrasG12D site mutation BxPC-3 cell line by CRISPR/Cas9
PCS-sgRNA (CRISPR/Cas9 vector) and TV-KrasG12D (Targeting vector) were designed and constructed by Biocytogen Co. Ltd (Beijing, China). BxPC-3 was transfected TV-KrasG12D and PCS-sgRNA by using the Neon transfection system (Life Technologies, Grand Island, NY). Transfected cells were selected in selective medium containing 200 ng/mL puromycin antibiotic for 3–5 days. Multiple monoclones were picked and cultured individually in separate wells.
Genotyping PCR
The puromycin-resistant clones were screened for homologous recombination by genomic PCR primers. The correct colony was detected Kras mutations by sequencing. The PCR fragments were directly sequenced or cloned into the pMD18-T (Takara, Japan) vector and then sequenced to identify Kras mutations.
Clonogenic Assay
Five hundred cells were seeded into each well of 6-well plate in triplicates, and colonies were stained 7–10 days later with 0.2% crystal violet in 80% methanol.
Anchorage-Independent Growth Assay
Ten thousand cells per well of 6-well plate were seeded in medium containing 1.6% Methyl cellulose (Sigma, M0512) with 10% fetal calf serum and plated over a layer of 0.9% agar-coated six-well plates. Standard medium (1 mL) was added to the top of the gelled matrix. After 15 days of culture, colonies were counted in five random three-dimensional fields per well.
Flow cytometry analysis of mitochondria mass and transmembrane potential
Cells were stained with 200 nM Mito Tracker Green (Invitrogen, Carlsbad, CA, USA) for 30 min to measure the mitochondrial mass and with 1 µM rhodamine-123 (30 min) to evaluate the mitochondrial transmembrane potential (MMP). Cells were washed with ice-cold phosphate-buffered saline (PBS) and kept on ice in the dark for immediate detection with a flow cytometry (FACS Aria II, BD Biosciences, CA, USA).
Immunofluorescence and confocal microscopy
Cells were cultured on sterilized glass slide covers until 70–80% confluence and incubated with 200 nM Mito Tracker Green for 30 min and with 5 µg/mL Hochest 33342 for 15 min. Then the cells were washed with PBS fixed with 3.7% paraformaldehyde. The slide covers were mounted on glass slides. Images were taken by LSM 710 confocal microscope (Carl Zeiss AG, Oberkochen, German).
Isolation of functional mitochondrial
The crude mitochondrial fractions were obtained using differential centrifugation from cultured cells as described previously with some modification (10,11). Cells were suspended in 1 mL IBc buffer (10 mM Tris-MOPS, 200 mM Sucrose, pH=7.4). The suspension was homogenized 30-gauge needle of syringe. Cells were lysed by forceful passage of the homogenate with 5–10 strokes. Lysis was monitored microscopically. After cell disruption, an aliquot of the lysate was examined to ensure that 80% of the cells were lysed. The homogenate was centrifuged at 600 g for 10 min at 4 °C. The supernatant was centrifuged at 7,000 g for 10 min at 4 °C. The pellet was collected and washed with IBc buffer once. The mitochondrial suspension is ready to be used for functional analysis.
Spectrofluorometry measurements of isolated mitochondrial mass
The cells were stained by 200 nM Mito Tracker Green for 30 min and mitochondrial was isolated. The experimental work was performed by exciting Mito Tracker Green at 480 nm and detecting the peak of fluorescence emission at 515 nm.
Western blot and qPCR
Active Ras protein was measured using RAS active detection kit (Cell signaling Technology, Danvers, MA, USA) according to instruction. For regular western blot, protein extracts were prepared with the ice-cold high efficiency RIPA lysis buffer (Beijing Solarbio Science & Technology Co., Ltd.). Twenty micrograms of each total protein were applied on 5–15% SDS-PAGE according to the molecular weight of detected proteins, then subjected to electrophoretic analysis and blotting. All primary antibodies (Cell Signaling Technology) were incubated overnight at 4 °C; and then incubated with peroxidase-coupled antibody at RT for 1 h which was used for detecting the primary antibody binding. Protein bands were visualized with an enhanced chemiluminescence kit (Merck Millipore).
For qPCR, cells were collected and total RNA was isolated with RNAiso Plus (Takara). qPCR was performed with SYBR Green (Takara). For COX6C mRNA expression, the primer sequences were: forward 5'-ctttgtataagtttcgtgtgg-3' and reverse 5'-attcatgtgtcatagttcagg-3'.
siRNA transfection
The COX6C specific siRNA (RiboBio, Guangzhou, China) was transfected with Lipofectamine RNAiMAX (Life Technologies) in six-well plates. The siRNA sequence against COX6C was sense 5'-GGACCACAUUAGGAAGGUUTT-3' and anti-sense 5'-AACCUUCCUAAUGUGGUCCAG-3'. The negative control siRNA was used as non-targeting control for all siRNA experiment. Following 48 h transfection, the cells subject to be detected by Western Blot or isolated mitochondrial.
ATP quantification
The detection of ATP from the isolated mitochondrial was determined based on luciferin/luciferase method with bioluminescent ATP determination assay kit (Molecular Probes, Bethesda, MD, USA) according to the manufacturer’s protocol
Gene array analysis
The human mitochondrial energy metabolism PCR array (Qiagen, #PAHS-008Z, Hilden, Germany) was used, which includes 84 key genes involved in mitochondrial respiration. Briefly, total RNA was extracted from cells and cDNA was mixed with the SYBR Green qPCR Master Mix and loaded into a 96-well plate for qPCR analyses performed on an ABI7500 Fast PCR machine (Applied Biosystems, Foster City, CA, USA) following the instructions provided by the PCR array kit. qPCR data was analyzed using online software (Qiagen).
Tissue microarray and immunohistochemistry staining
The tissue microarray chips containing a total of 30 pairs of pancreatic carcinoma and matched adjacent tissues were provided by Shanghai Outdo Biotech Co., Ltd. (Shanghai, China). Briefly, the main experimental procedure is as follows: sections were proceeded as dewaxing, microwave antigen retrieval, endogenous peroxidase blocking, and then incubated with the COX6C antibody (TA506178, 1:100, OriGene, Rockville, MD, USA) overnight, and the second antibody for 0.5 h, respectively. Finally, the specimens were determined with DAB detection and hematoxylin staining.
COX enzyme assays
Isolated mitochondrial fraction was resuspended in 20 volumes of ice-cold resuspended buffer (25 mM potassium phosphate, 5 mM MgCl2, 1 mM EDTA, 0.6 mM lauryl maltoside) with protease inhibitors. Samples were freeze-thawed three times. Protein concentration was determined according BCA kit. Homogenates were diluted 10× assay buffer and added to 96-well plate containing assay buffer and reduced 0.05 mM cytochrome c. Absorbance at 550 nm was followed for 30 min in a spectrophotometer (Beckman, Brea, CA, USA).
Statistical analysis
Results were expressed as mean ± SD. Comparisons were made between different treatments using unpaired Student’s t-test.
Results
Fast proliferation characterization of BxPc-3/KrasG12D cells
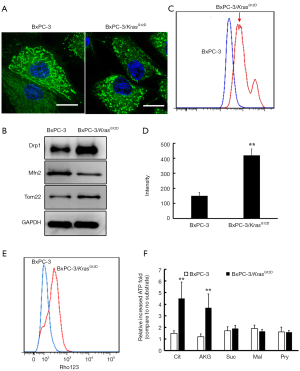
We generated a transgenic cell line which was induced KrasG12D by taking advantage of CRISPR/Cas9 genome modification in Kras wild type cells (BxPC-3). In this new cell line-BxPC-3/KrasG12D, G12D (12th amino acid glycine in Kras gene replaced with aspartate) was introduced to exon 1 locus of endogenous Kras gene and therefore was expressed under the control of Kras promoter. By inserting KrasG12D into downstream of starting codon in BxPC-3 cells Kras locus, we expect KrasG12D to be expressed under the endogenous Kras transcriptional control. Briefly, a pair of vectors including sgRNA vector and knockin KrasG12D vector were mixed and electro-transfected into BxPC-3 cell. Anti-neomycin colonies were expanded. Subsequently genotyping PCR screening was to confirm the homologous recombination and include KrasG12D mutation site. The correct colonies were established and named as BxPC-3/KrasG12D cells. Activated Kras expression of BxPC-3/KrasG12D cells was detected by the GTP-bound GTPase pull down assay. In Figure 1A, activated Kras expression in BxPC-3/KrasG12D cells was higher than wild type BxPC-3. This indicated the Kras activity in BxPC-3/KrasG12D cells was increased by Kras mutation. BxPC-3/KrasG12D cells exhibit enhanced proliferative properties compared with wild type BxPC-3 by using colony formation assay (Figure 1B). Next, the anchorage-independent growth both of cells were examined using the soft agar assay. We observed that the activation of KrasG12D expression enabled an elevated anchorage independent growth capability of BxPC-3/KrasG12D cells compared with BxPC-3 (Figure 1C).

Mitochondrial characterizations of BxPc-3/KrasG12D cells
The fluorescence microscopy was used to observe the mitochondrial morphology. The wild type BxPC-3 cells staining with Mito Tracker Green were shown network connection. However, BxPC-3/KrasG12D cells were seen more disconnected (more dot-shaped) mitochondrial (Figure 2A). Western blot analysis showed that Drp-1 was up-regulated and Mfn-2 was down-regulated in BxPC-3/KrasG12D cells compared with the wild type BxPC-3 (Figure 2B). We further characterized mitochondrial mass in BxPC-3/KrasG12D cells. Fluorescence intensity of cells staining Mito Tracker Green was detected by flow cytometry. The mitochondrial mass in BxPC-3/KrasG12D cells was moderately increased (Figure 2C). The isolated mitochondrial labeled Mito Tracker Green was detected by spectrofluorometer. The fluorescence intensity of mitochondrial in BxPC-3/KrasG12D cells was much higher than that in wild type BxPC-3 (Figure 2D). Western blot analysis showed that overexpression of Tom22, a core component of the mitochondria outer membrane protein translocation pore, was up-regulated in BxPC-3/KrasG12D cells (Figure 2B). All these data showed that the mitochondrial mass was increased after KrasG12D activation. Mitochondrial membrane potential of cells staining with Rhodamine 123 was estimation by flow cytometry. BxPC-3/KrasG12D cells showed higher mitochondrial membrane potential than wild type BxPC-3 cells (Figure 2E). ATP production was estimated by isolated mitochondrial from both of the cells. ATP content showed similar basal levels in both cells, whereas which was increased after adding TCA metabolites. This indicated the isolated mitochondrial from the cells kept intact functional. The increased ATP content was average around 2 times after adding TCA cycle intermediates include citrate (Cit), α-ketoglutarate (AKG), succinate (Suc), malate (Mal) and pyruvate (Pyr) compared with basal levels in wild type BxPC-3 cells (Figure 2F). However, ATP of isolated mitochondrial in BxPC-3/KrasG12D cells was remarkably increased when AKG and Cit treatment. 4.5 folds were increased treated with AKG and 3.7 folds were increased treated with Cit.
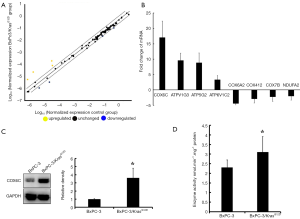
Expression and activities of COX6C in BxPC-3/KrasG12D cells
We employed the human mitochondrial energy metabolism PCR array to screen the differential energy metabolism related genes from the pairs of cells. Figure 3A is the scatter plot which compared the normalized expression of each gene on the array between two group cells (BxPC-3/KrasG12D cells vs. wild type BxPC-3 cells). The central line indicated unchanged gene expression and the dotted lines indicated the 2-fold regulation threshold. Based on gene selection criteria (P<0.05 and fold change ≥2), there were 4 of 84 genes showed up-regulated expression and 4 genes showed down-regulated expression in BxPC-3/KrasG12D cells compared with wild type BxPC-3 (Figure 3B). Gene expression profiles differed maximum between two cells was COX6C gene. The relative expression ratio of the mRNA of COX6C gene in both cells was analyzed by RT-PCR. The mRNA expression of COX6C in BxPC-3/KrasG12D cells (18.2-fold elevation) was higher than in wild type BxPC-3 cells. COX6C was up-regulated expression confirmed by Western Blot (Figure 3C). The protein expression was found to be elevated 4.6-fold in BxPC-3/KrasG12D cells compared with wild type BxPC-3. We then detected the activities of cytochrome c oxidase. As shown in Figure 3D, mean enzymatic activity was increased by 35% in BxPC-3/KrasG12D cells compared with wild type BxPC-3.
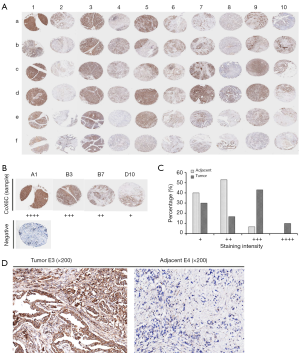
The expression level of COX6C via tissue microarray and immunohistochemistry
On the basis of the above gene microarray, we tested expression of COX6C through tissue microarray of pancreatic carcinoma and matched adjacent tissues. The scanning pattern image of COX6C was presented in Figure 4A. The evaluation standards were presented in Figure 4B. According to the evaluation standards, the classification of samples was shown in Figure 4C. Weak cytoplasmic COX6C IHC staining were shown in adjacent tissue group, and moderate to strong staining cytoplasmic COX6C IHC staining were shown in tumor group. COX6C immunoreactivity was significantly more intense and diffuse in tumor tissue compared with matched adjacent tissue. As shown in Figure 4D, the representative results between tumors and their matched adjacent tissues denoted significant difference. Evidently, it suggested that significant COX6C overexpression was identified in pancreatic cancer tissue compared with matched adjacent tissue.
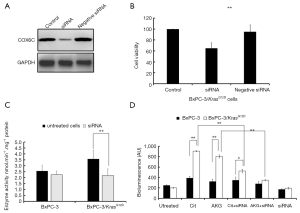
Effect of COX6C siRNA on ATP production in BxPC-3/KrasG12D cells
To investigate whether silencing COX6C expression could affect the metabolite characterization of cells, BxPC-3/KrasG12D cells were transfected with COX6C siRNA for 48 h. As shown in Figure 5A, COX6C siRNA transfection decreased significantly COX6C levels compared with the negative siRNA control. Cells viability was inhibited by COX6C siRNA compared with the negative siRNA control. (Figure 5B). Enzyme activity of COX decreased significantly in COX 6C siRNA transfected BxPC-3/KrasG12D cells (Figure 5C). We further investigated whether silencing COX6C expression could affect the ATP production of mitochondrial. Both of cells were transfected with COX6C siRNA for 48 h, mitochondria were isolated and incubated with 10 mM different substrates for 15 min at 37 °C in dark 96-well plates. The amount of ATP generated from mitochondria was quantified using ATP determination kit. As shown in Figure 5D, there are no statistical significance in endogenous mitochondria BxPC-3/KrasG12D cells compared with wild type BxPC-3 cells (ATP generation from residual endogenous substrates, no exogenous substrate added). However, when treated with Cit or AKG, mitochondria isolated from BxPC-3/KrasG12D cells produced a significant amount of ATP increase. When transfected with COX6C siRNA, there was a significant decrease of ATP production in BxPC-3/KrasG12D cells and slightly decrease in wild type BxPC-3 cells.
Discussion
There are newer reviews on the contribution of Ras mutations and altered Ras signaling in cancers, and in pancreatic cancer in particular (12). There are still no specific therapies for directly targeting mutant RAS protein currently available in clinics (13). Targeting Ras downstream pathway is currently strategy. Kras mutation has been linked to many alterations of metabolism-related characteristics. These metabolic adaptations including the Warburg effect have been recognized as one of the hallmarks of cancer (14). Taken together, the alteration of metabolic adaptations of Ras-mutant cancer cell may provide novel therapeutic opportunities.
Normally, different Kras mutation cells lines are independently derived cell lines that differ in many phenotypic and genetic properties. In this study, by taking advantage of the CRISPR/Cas9 system, we generated a KrasG12D knockin cell line. Compared with wild type BxPC-3, BxPC-3/KrasG12D maintained maximum genetic similarity besides the specific Kras mutation site, which allowed us to explore the precise alteration driven by Kras mutation.
It is known mitochondria is the highly dynamic organelle that constantly changes shape and structure in response to different metabolic demands of the cells. BxPC-3/KrasG12D cells led to rapid mitochondrial division, and expression of Drp1 compared with the wild type BxPC-3, which indicates KrasG12D activation leads to going through more mitochondrial division.
We searched the differential genes between the wild type BxPC-3 and BxPC-3/KrasG12D cells by screening human mitochondrial energy metabolism PCR array. In all the tested 84 key genes involved in mitochondrial respiration, the COX6C is the highest overexpressed gene driven in BxPC-3/KrasG12D cells. COX, as one of the electron-driven proton pumps of oxidative phosphorylation, plays a critical role in energy metabolism (15,16). COX is encoded by two genomes: 10 nuclear-encoded subunits and three subunits encoded by mitochondrial DNA (17,18). Alteration a single enzyme complex (COX) is commensurate with tumor-altered metabolism (19,20). Varieties of COX subunits have been connected with cancer. The expression of cytochrome c oxidase subunit is overexpressed in colon cancer (21,22). The COX5A play a role in invasion of lung cancer (23). Functional effects of altered cytochrome c oxidase are related to prostate cancer (24,25).
In our study, COX6C is found not only the highest up-expression gene among all the 84 detected mitochondrial respiration related genes, but also the only up-expression gene among all the 14 of COX genes. COX6C as one of the subunits of COX holoenzyme is encoded by a nuclear gene. COX6C protein involves in the respiration chain in the mitochondrial inner member. Enhanced expressions of COX6C in prostate carcinoma cancer have been described (26). However, limited literature has been found to report its biology function with cancer. Here, we first provided evidence that COX6C was up-regulated expression in both mRNA and protein level in KrasG12D driven cells. Similar result of linkage COX and Ras mutation has also been reported that activation of HRas (V12) increases COX activity and mitochondrial respiration in part via up-regulation of COX Vb (20). The high expression of COX6C in pancreatic cancer tissue compared with that adjacent normal tissue provided strong evidence allowing us to determine its value as a marker related to energy metabolite.
COX6C knockdown significantly decreased ATP production treated with Cit (60% decrease) and with AKG (40% decrease) in isolated mitochondrial from BxPC-3/KrasG12D cells. Although the expression levels of COX subunits differ between two cells, the assays for COX activity we employed reflect differences in COX holoenzyme levels between two cell lines. The other subunits involved in COX activity detected in our PCR array, 3 of them are down-regulated and other genes weren’t affected in BxPC-3/KrasG12D cells. These findings suggested that expression of COX6C could play a crucial role in elevated COX activity and ATP production in BxPC-3/KrasG12D cells.
Our studies carry out that activation of KrasG12D in pancreatic cancer cells increases the COX activity and ATP production of mitochondrial via up-regulation of COX6C. These results indicate this regulatory subunit COX6C may have utility as a Ras effector target for development of anti-pancreatic cancer therapeutics, especially against tumors in which the RasG12D driven singling pathway is activated.
Acknowledgments
Funding: This work was supported by Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Sciences (2017-I2M-1-010) and by Natural Science Foundation of China [81771860].
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.03.02). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lv J, Wang J, Chang S, et al. The greedy nature of mutant RAS: a boon for drug discovery targeting cancer metabolism. Acta Biochim Biophys Sin (Shanghai) 2016;48:17-26. [PubMed]
- Kanda M, Matthaei H, Wu J, et al. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology 2012;142:730-3. [Crossref] [PubMed]
- Bryant KL, Mancias JD, Kimmelman AC, et al. KRAS: feeding pancreatic cancer proliferation. Trends Biochem Sci 2014;39:91-100. [Crossref] [PubMed]
- Ying H, Kimmelman AC, Lyssiotis CA, et al. Oncogenic KRAS maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 2012;149:656-70. [Crossref] [PubMed]
- Son J, Lyssiotis CA, Ying H, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 2013;496:101-5. [Crossref] [PubMed]
- Lock R, Roy S, Kenific CM, et al. Autophagy facilitates glycolysis during RAS-mediated oncogenic transformation. Mol Biol Cell 2011;22:165-78. [Crossref] [PubMed]
- Commisso C, Davidson SM, Soydaner-Azeloglu RG, et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 2013;497:633-7. [Crossref] [PubMed]
- Wang W, Fang G, Rudolph J. Ras inhibition via direct Ras binding—is there a path forward. Bioorg Med Chem Lett 2012;22:5766-76. [Crossref] [PubMed]
- Cox AD, Fesik SW, Kimmelman AC, et al. Drugging the undruggable RAS: Mission Possible. Nat Rev Drug Discov 2014;13:828-51. [PubMed]
- Frezza C, Cipolat S, Scorrano L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat Protoc 2007;2:287-95. [Crossref] [PubMed]
- Hornig-Do HT, Günther G, Bust M, et al. Isolation of functional pure mitochondria by superparamagnetic microbeads. Anal Biochem 2009;389:1-5. [Crossref] [PubMed]
- Fernández-Medarde A, Santos E. Ras in Cancer and Developmental Diseases. Genes Cancer 2011;2:344-58. [Crossref] [PubMed]
- Baker NM, Der CJ. Cancer: Drug for an 'undruggable' protein. Nature 2013;497:577-8. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Villani G, Attardi G. In vivo control of respiration by cytochrome c oxidase in human cells. Free Radic Biol Med 2000;29:202-10. [Crossref] [PubMed]
- Srinivasan S, Avadhani NG. Cytochrome c oxidase dysfunction in oxidative stress. Free Radic Biol Med 2012;53:1252-63. [Crossref] [PubMed]
- Grossman LI, Lomax MI. Nuclear genes for cytochrome c oxidase. Biochim Biophys Acta 1997;1352:174-92. [Crossref] [PubMed]
- Dhar SS, Ongwijitwat S, Wong-Riley MT. Chromosome conformation capture of all 13 genomic loci in the transcriptional regulation of the multisubunit bigenomic cytochrome c oxidase in neurons. J Biol Chem 2009;284:18644-50. [Crossref] [PubMed]
- Krieg RC, Knuechel R, Schiffmann E, et al. Mitochondrial proteome: Cancer-altered metabolism associated with cytochrome c oxidase subunit level variation. Proteomics 2004;4:2789-95. [Crossref] [PubMed]
- Telang S, Nelson KK, Siow DL, et al. Cytochrome c oxidase is activated by the oncoprotein Ras and is required for A549 lung adenocarcinoma growth. Mol Cancer 2012;11:60-5. [Crossref] [PubMed]
- Payne CM, Holubec H, Bernstein C, et al. Crypt-restricted loss and decreased protein expression of cytochrome c oxidase subunit I as potential hypothesis-driven biomarkers of colon cancer risk. Cancer Epidemiol Biomarkers Prev 2005;14:2066-75. [Crossref] [PubMed]
- Zhang K, Chen Y, Huang X, et al. Expression and clinical significance of cytochrome c oxidase subunit IV in colorectal cancer patients. Arch Med Sci 2016;12:68-77. [Crossref] [PubMed]
- Chen WL, Kuo KT, Chou TY, et al. The role of cytochrome c oxidase subunit Va in non-small cell lung carcinoma cells: association with migration, invasion and prediction of distant metastasis. BMC Cancer 2012;12:273-8. [Crossref] [PubMed]
- Namslauer I, Dietz MS, Brzezinski P. Functional effects of mutations in cytochrome c oxidase related to prostate cancer. Biochim Biophys Acta 2011;1807:1336-41. [Crossref] [PubMed]
- Herrmann PC, Gillespie JW, Charboneau L, et al. Mitochondrial proteome: Altered cytochrome c oxidase subunit levels in prostate cancer. Proteomics 2003;3:1801-10. [Crossref] [PubMed]
- Wang FL, Wang Y, Wong WK, et al. Two differentially expressed genes in normal human prostate tissue and in carcinoma1. Can Res 1996;56:3664-7.


