
ZATT, TDP2, and SUMO2: breaking the tie that binds TOP2 to DNA
Type II topoisomerase (TOP2) poisons are widely used chemotherapeutics that work by disrupting the cycle of TOP2-DNA interactions that are required for DNA synthesis, gene expression, and the maintenance of genome integrity. Schellenberg et al.’s recent Science paper, entitled “ZATT (ZNF451)-mediated resolution of topoisomerase 2 DNA-protein cross-links” (1) provides a major advancement in the understanding of the mechanisms that regulate TOP2-DNA interactions, and how they can inhibit the anticancer effects of TOP2 poisons. Through these new insights, promising new therapeutic targets have been identified.
Topoisomerases II-α and II-β (TOP2-α and TOP2-β) enzymes are vital in the unwinding of DNA during cell processes such as transcription, DNA synthesis, and mitosis. TOP2s create and repair DNA double strand breaks in duplex DNA, allowing one strand to move through the other, thus preventing DNA supercoiling (2) (Figure 1A). Critical in the function of TOP2s is the formation of a transient intermediate, called the DNA TOP2 cleavage complex (TOP2cc). When functioning normally, this TOP2cc is short-lived. However, in cells treated with a class of anticancer compounds called TOP2 poisons, the TOP2cc becomes more stable and is “trapped” in this conformation (3). Trapped TOP2cc can then be degraded in the cell via proteolysis, releasing the broken DNA inside (4) (Figure 1B). These DNA double strand breaks are typically cell lethal. Since TOP2 activity is particularly active in rapidly proliferating cells, TOP2 poisons can therefore be quite effective against cancerous cells (3).
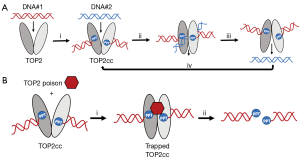
Cells also possess tyrosyl-DNA phosphodiesterase 2 (TDP2), a protein involved in the resolution of TOP2-DNA linkages. Through this function, TDP2 can help cells overcome the deleterious effects of TOP2 inhibition or poisoning (5,6). It is therefore a promising target for novel chemotherapies (7,8). However, an important question remains; how does TDP2 access and hydrolyze the protected TOP2-DNA phosphotyrosyl bond to facilitate TOP2cc repair? Schellenberg et al. sought to answer this question.
To identify modulators of TDP2-dependent repair of TOP2cc, the authors used yellow-fluorescent protein (YFP)-tagged TDP2 stably expressed in human embryonic kidney (HEK) 293F cells, which allowed for the collection and analysis of immunoprecipitated TDP2-containing protein complexes. These experiments confirmed the known interaction between TOP2 and TDP2. Interestingly, there was enrichment of a protein called small ubiquitin-related modifier 2 (SUMO2). Given that SUMOylation, the covalent attachment of SUMO protein to lysine residues in a target protein (9), promotes protein-protein interactions during the DNA damage response (10), Schellenberg et al. identified SUMO2 as a protein of interest in the resolution of TOP2-DNA crosslinks. Their subsequent experiments showed that TDP2 formed bonds with intact, non-proteolyzed TOP2-α and TOP2-β, which is then SUMOylated and subsequently liberated from the chromatin. These interactions were abolished when a catalytically inactive variant of TDP2 (labelled TDP2H35IN) was used. These results suggested that, contrary to the prevailing hypothesis, TDP2 processes TOP2-DNA oligopeptides independently of proteasome degradation of TOP2cc. Supporting the above results, TDP2 repaired etoposide-induced DNA double strand breaks in mouse embryonic fibroblast (MEF) cells in the presence of the proteasome inhibitor, MG132. Together, these experiments help clarify that this mechanism of TOP2cc resolution involves both TDP2 and SUMO2, two proteins that act independently of, or parallel to, proteasome-mediated TOP2cc repair.
However, the regulatory pieces of the puzzle connecting SUMO2 and TDP2 in the process of TOP2cc resolution were still missing. To resolve this, Schellenberg et al. used an elegant procedure called tandem affinity purification (TAP). Briefly, the authors expressed YFP-TDP2 and His6-SUMO2 in HEK293F cells. Using an anti-GFP sephadex, which binds to YFP, they pulled down the YFP-TDP2 associated proteins, which would include His6-SUMO2. Then, a Ni-nitrilotriacetic acid (NTA) agarose column, which recognizes His6-SUMO2, was used to pull out the His6-SUMOylated proteins. This isolate was then digested with the SUMO-protease, ubiquitin-like specific protease 1, leaving behind any proteins that were bonded to the SUMO2. Finally, the identities of the interacting proteins were determined using LC/MS-MS. The strength of this method is that it was able to identify proteins that were connected to both TDP2 and SUMO2. As expected, TOP2α and TOP2β were identified as factors involved in SUMO2- and TDP2-dependent TOP2cc repair. Of particular interest, the TAP assays identified an additional interacting protein, zinc finger protein 451 (ZNF451), a SUMO2 E3/E4 ligase that may have formed a functional complex with SUMOylated TOP2 and TDP2 within the cells. Schellenberg et al. determined that ZNF451 actually bonds to TOP2α and TOP2β and is recruited to the cellular chromatin fraction after etoposide poisoning of TOP2, suggesting a putative role of ZNF451 in proteasome-independent resolution of TOP2cc repair. To test this idea, a reconstituted TOP2cc was generated which could be assayed for TDP2-dependent resolution. The resolution of TOP2cc by TDP2 was more than three-fold higher in the presence of ZNF451 compared to TDP2 alone, and neither ZNF451 alone or inactive TDP2H35IN catalyzed this reaction. This data suggested that ZNF451 stimulates TOP2cc hydrolysis catalyzed by TDP2. In HEK293 cells, knockdown of ZNF451 prevented the resolution of etoposide-induced DNA double strand breaks whereas overexpression of ZNF451 had the opposite effect. The inhibition of DNA damage resolution was even more pronounced when ZNF451 knockdown was combined with TDP2 knockdown. Based on these results, it can be concluded that ZNF451 is an important component in the cellular response to TOP2 poison-induced DNA damage, and that it operates both independently and in conjunction with TDP2.
With evidence pointing to the importance of ZNF451, a SUMO2 E3/E4 ligase (11), the next logical step was to determine if ZNF451-mediated TOP2 SUMOylation facilitates resolution of the TOP2cc. ZNF451 displayed a significant preference for the SUMOylation of TOP2cc over free TOP2, suggesting TOP2cc is a specific target of ZNF451 SUMO2 ligase activity. Likewise, in HEK293F cells, etoposide-induced SUMOylation of TOP2 was heavily dependent on ZNF451, supporting that ZNF451 induces the SUMOylation of TOP2cc in cells after they are treated with TOP2 poisons. Next, “in vivo complexes of enzymes” (ICE) assays were used to monitor TOP2β and SUMO2-modified TOP2β DNA-protein cross-link removal from chromatin after etoposide poisoning and removal. While proteasome inhibition reduced the rate at which SUMO2-modified TOP2β was removed from the chromatin, there was no effect of TDP2 knockdown in the presence or absence of proteasome inhibition. This result was somewhat at odds with the majority of other data supporting that the SUMOylation of TOP2cc catalyzes TDP2-dependent TOP2cc resolution. The exact reason for this discrepancy is unclear.
Using maltose binding protein pulldowns to identify protein-binding partners, the authors then determined that SUMO2 binds to the catalytic domain of TDP2 (labelled TDP2cat). This was unexpected since the catalytic domain lacks a standard SUMO interaction motif (SIM) (12). They then used X-ray crystallography to determine the structure of mouse TDP2cat-SUMO2 and TDP2cat-SUMO2-DNA complexes. Of particular interest was the discovery that the core of the TDP2-SUMO2 interface is composed of a “split-SIM” structure, meaning that TDP2 interacts with SUMO2 at two distinct points rather than the usual one. This split-SIM structure results in a particularly strong bond between the two proteins, placing TDP2 among the stronger of the known SUMO-binding proteins (12). By synthesizing mutant versions of TDP2 (TDP2C-AE), they determined the importance of these TDP2-SUMO2 interface points as all mutants failed to colocalize with SUMO2. Then, when ZNF451 was knocked down, resulting in a lack of TOP2 SUMOylation, the interactions between TDP2 and TOP2 were again impaired. Together, these data revealed the importance of the TDP2-SUMO2 interface in its interactions with SUMOylated TOP2. To explore the kinetics of this reaction, TDP2 recruitment to TOP2 was measured following DNA damage by UV radiation. ZNF451 knockdown and mutant TDP2C-AE impaired TDP2 accumulation at the DNA damage site. Furthermore, the mutant TDP2C-AE delayed the removal of the SUMOylated fraction of TOP2 from the previously purified TOP2cc. Together, these results demonstrated that the TDP2-TOP2cc interaction is enhanced by TDP2-SUMO interactions, and that the TDP2-TOP2cc interaction is dependent on the SUMO2 ligase, ZNF451.
Overall, Schellenberg et al. concluded that the ZNF451-TDP2-modulated TOP2cc resolution pathway (Figure 2) could contribute to the development of drug resistance in cancer cells to TOP2 poisons, suggesting a novel pathway that could be targeted to improve anticancer therapies. While ZNF451 was shown to be heavily involved in the activity of TDP2, the authors also found evidence of TDP2-independent effects of ZNF451 on the cellular response to TOP2 poisoning that they have yet to explain, warranting further research. It will also be important to better understand the exact mechanics through which ZNF451-TDP2 influences TOP2, and ZNF451-TDP2’s possible modulation of TOP2 genomic and transcriptional regulation. Ultimately, Schellenberg et al. finished their article by suggesting a new name for ZNF451, based on its close association with TDP2-mediated TOP2 resolution and repair: ZNF associated with TDP2 and TOP2, or ZATT.
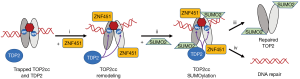
TDP2 was previously shown to be involved in cell survival after treatment with TOP2 poisons (5,6). Schellenberg et al.’s research provides a clearer picture of exactly how TDP2 associates with TOP2 when resolving TOP2cc, how its activity helps increase cell survivability when being treated with TOP2 poisons, and the extent to which SUMOs and the SUMO2 ligase ZNF451 are involved in this TOP2cc resolution mechanism. Through these novel findings, the authors have advanced the field of topoisomerase research by improving the understanding of TOP2cc resolution; a fundamental step in the TOP2 DNA-cleaving and re-ligating process.
The authors’ research findings are particularly important for cancers that have become resistant to TOP2 poisons. TOP2 poison resistance can develop in a variety of different ways, such as site-specific mutations on TOP2 proteins, alterations in TOP2 phosphorylation (14), and enhanced drug efflux (15). The authors have uncovered a potential new avenue through which resistance to TOP2 poisons can occur in cancer cells for which there are both negative and positive implications. The downside is that the overexpression or mutation of ZNF451 and/or TDP2 could result in enhanced activity of the ZNF451-TDP2-modulated TOP2cc resolution pathway during chemotherapy with TOP2 poisons, improving cancer cell survivability. The upside is that, by uncovering this mechanism, new putative targets for chemotherapeutic intervention have been identified. Specifically, the inhibition of ZNF451 and/or TDP2 should help prevent TOP2cc from resolving after TOP2 poisoning, which could significantly improve the efficacy of TOP2 poisons in both drug-naïve and multidrug resistant cancer cells. In support of this hypothesis, a small molecule inhibitor of TDP2 has been shown to enhance etoposide activity in lung cancer cells that highly express TDP2 (16). In the same study, TDP2 was found to be highly expressed in the majority of lung and breast tumour biopsies from patients, suggesting its inhibition could prove to be an effective treatment clinically (16).
Through their very thorough experiments, Schellenberg et al. also convincingly showed that TDP2 and ZNF451 can process TOP2-DNA oligopeptides without the help of proteolytic degradation. Based on the prevailing hypothesis prior to their work, one may have predicted that co-treatment with a proteasome inhibitor would be a sure-fire method to increase the efficacy of TOP2 poisons. Some studies support this idea. For example, in Lee et al. the proteasome inhibitor MG132 potentiated the activity of a variety of TOP2 poisons in vitro (17). However, the work of Schellenberg et al. strongly suggests that the unencumbered activity of TDP2 and ZNF451 would allow for a significant amount of TOP2cc to be resolved anyway, even with proteasomal inhibition. Therefore, inhibitors of TDP2 and ZNF451 may prove to be more efficacious combination therapies with TOP2 poisons, versus proteasome inhibitors.
While this is a very thorough and convincing study, there are certain limitations to note and additional questions that emerge from the results. The first limitation is that the cellular studies were restricted to healthy HEK 293F and MEF cells. An important next step will be to more broadly assess the function of ZNF451 and TDP2 in cancers that would normally be treated with topoisomerase II poisons, such as metastatic breast cancer or leukemia. For example, does TDP2 and ZNF451 expression predict cancer cell sensitivity to TOP2 poisons? Or can ZNF451 and TDP2 mutations exist in cancer cells, and would such mutations provide a survival advantage and innate resistance to drugs that target TOP2? Second, while ZNF451-TDP2-modulated TOP2cc resolution represents a novel pathway for possible chemotherapeutic intervention, this was not tested experimentally in their study. However, there is at least one study showing that small molecule inhibition of TDP2 enhances the efficacy of TOP2 poisons (16). Given the authors’ findings, there is likely to be considerable interest in the coming years to identify small molecule inhibitors of ZNF451. As such compounds are developed, it will allow for researchers to address the critical questions: does ZNF451 and/or TDP2 inhibition alone have anticancer effects and, when given in combination with TOP2 poisons, do such inhibitors significantly improve their anticancer activity in drug-resistant cancer cells? Third, the studies only used a single TOP2 poison, etoposide, which is a non-intercalating poison. It will be important to validate the authors’ findings using intercalating TOP2 poisons, such as doxorubicin or mitoxantrone (18), to determine if the mechanisms of TOP2cc resolution are conserved across sub-classes of TOP2 poisons. Fourth, one must be wary of the potential for off target or adverse drug effects that could stem from inhibition of ZNF451 and TDP2 activity. For example, the TOP2 poison, doxorubicin, which is a very effective chemotherapeutic, is also quite cardiotoxic. Doxorubicin targeting of TOP2β, which triggers TOP2β degradation by proteasomes and DNA damage in cardiac cells, may contribute to this adverse effect (19). If inhibitors of ZNF451 and TDP2 can increase DNA damage caused by TOP2β poisons, it is conceivable that they may also increase their cardiotoxic properties. Therefore, future studies exploring the putative side effect profiles of ZNF451 and TDP2 inhibition will be needed.
In summary, through a comprehensive investigation, Schellenberg et al. have identified an exciting new TOP2cc resolution pathway involving the direct action of ZNF451 (or ZATT) and TDP2 which functions independently of proteasome activity. These findings open up new possibilities for cancer treatments, especially in cancers that have developed resistance to current TOP2 poisoning chemotherapeutics.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Yan Li (Experimental Therapeutics Centre, Agency for Science, Technology and Research, Singapore).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.02.03). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Schellenberg MJ, Lieberman JA, Herrero-Ruiz A, et al. ZATT (ZNF451)-mediated resolution of topoisomerase 2 DNA-protein cross-links. Science 2017;357:1412-16. [Crossref] [PubMed]
- Pommier Y, Leo E, Zhang H, et al. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol 2010;17:421-33. [Crossref] [PubMed]
- Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer 2009;9:338-50. [Crossref] [PubMed]
- Mao Y, Desai SD, Ting CY, et al. 26 S proteasome-mediated degradation of topoisomerase II cleavable complexes. J Biol Chem 2001;276:40652-8. [Crossref] [PubMed]
- Schellenberg MJ, Perera L, Strom CN, et al. Reversal of DNA damage induced Topoisomerase 2 DNA-protein crosslinks by Tdp2. Nucleic Acids Res 2016;44:3829-44. [Crossref] [PubMed]
- Cortes Ledesma F, El Khamisy SF, Zuma MC, et al. A human 5'-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature 2009;461:674-8. [Crossref] [PubMed]
- Gómez-Herreros F, Schuurs-Hoeijmakers JH, McCormack M, et al. TDP2 protects transcription from abortive topoisomerase activity and is required for normal neural function. Nat Genet 2014;46:516-21. [Crossref] [PubMed]
- Hornyak P, Askwith T, Walker S, et al. Mode of action of DNA-competitive small molecule inhibitors of tyrosyl DNA phosphodiesterase 2. Biochem J 2016;473:1869-79. [Crossref] [PubMed]
- Wilkinson KA, Henley JM. Mechanisms, regulation and consequences of protein SUMOylation. Biochem J 2010;428:133-45. [Crossref] [PubMed]
- Jackson SP, Durocher D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol Cell 2013;49:795-807. [Crossref] [PubMed]
- Eisenhardt N, Chaugule VK, Koidl S, et al. A new vertebrate SUMO enzyme family reveals insights into SUMO-chain assembly. Nat Struct Mol Biol 2015;22:959-67. [Crossref] [PubMed]
- Hecker CM, Rabiller M, Haglund K, et al. Specification of SUMO1- and SUMO2-interacting motifs. J Biol Chem 2006;281:16117-27. [Crossref] [PubMed]
- Zagnoli-Vieira G, Caldecott KW. TDP2, TOP2, and SUMO: what is ZATT about? Cell Res 2017;27:1405-6. [Crossref] [PubMed]
- Ganapathi RN, Ganapathi MK. Mechanisms regulating resistance to inhibitors of topoisomerase II. Front Pharmacol 2013;4:89. [Crossref] [PubMed]
- Hall SR, Toulany J, Bennett LG, et al. Jadomycins Inhibit Type II Topoisomerases and Promote DNA Damage and Apoptosis in Multidrug-Resistant Triple-Negative Breast Cancer Cells. J Pharmacol Exp Ther 2017;363:196-210. [Crossref] [PubMed]
- Kont YS, Dutta A, Mallisetty A, et al. Depletion of tyrosyl DNA phosphodiesterase 2 activity enhances etoposide-mediated double-strand break formation and cell killing. DNA Repair (Amst) 2016;43:38-47. [Crossref] [PubMed]
- Lee KC, Bramley RL, Cowell IG, et al. Proteasomal inhibition potentiates drugs targeting DNA topoisomerase II. Biochem Pharmacol 2016;103:29-39. [Crossref] [PubMed]
- Hasinoff BB, Wu X, Patel D, et al. Mechanisms of Action and Reduced Cardiotoxicity of Pixantrone; a Topoisomerase II Targeting Agent with Cellular Selectivity for the Topoisomerase IIα Isoform. J Pharmacol Exp Ther 2016;356:397-409. [Crossref] [PubMed]
- Lyu YL, Kerrigan JE, Lin CP, et al. Topoisomerase IIbeta mediated DNA double-strand breaks: implications in doxorubicin cardiotoxicity and prevention by dexrazoxane. Cancer Res 2007;67:8839-46. [Crossref] [PubMed]


