
WASF3 is associated with tumour invasiveness and confers a poor prognosis in human gastric cancer
Introduction
Gastric cancer (GC) is one of the most prevalent cancers in China and approximately half of GC patients are diagnosed at an advanced stage (1,2). However, distant metastasis (liver, peritoneal, lymph node metastasis and etc.) and recurrence after operation might be the main cause of death associated with GC (3,4). Clinical staging and histopathological criteria are the main methods to evaluate the prognosis of GC patients. The patients with advanced stage have poor prognosis under the active treatment (5). Therefore, identification of progression related molecules can help us find clues for accurate prognosis assessment, effective individualized treatment and improvement patient outcome.
WAS protein family member 3 (WASF3) is a member of the Wiskott-Aldrich syndrome protein family (6), which can form a multiprotein complex linking actin and receptor kinases, including ABI2, HSPC300/BRICK1, and CYFIP1-NCKAP1 proteins, etc. (7-11). Under stimulation of growth factors such as platelet-derived growth factor (PDGF), it can play a role in regulation of cell shape, motility and cytoskeletal organization through activating the Arp2/3 complex of proteins (12). Previous study indicated WASF3 levels are higher in advanced stage breast cancers compared to lower grade one and normal tissues (13). However, recently other studies have reported that WASF3 might play diverse roles in prostate cancer, non-small cell lung cancer, breast cancer and colorectal cancer (13-17).
Recent studies have showed that over-expression of WASF3 could increase the proliferation and movement of GC cells in vitro (18,19). However, little knowledge is known about the relationship between WASF3 expression and GC progression. In this study, we analysed WASF3 expression in GC by immunohistochemistry (IHC) to evaluated its correlation with clinicopathological features, and furthermore explore its possible function through knocking out its expression in vitro.
Methods
Cell culture and small guide RNA (sgRNA) plasmids
GC cell line was obtained from the Cell Research Institute (Shanghai, China). All cells were maintained in Dulbecco’s Modified Eagle’s medium (DMEM) (GIBCO BRL, Carlsbad, CA, USA) with supplementing 10% fetal calf serum (FCS) (GIBICO, Carlsbad, CA, USA), and penicillin/streptomycin in humidified atmosphere of 5% CO2 at 37 °C. CRISPR/Cas9 plasmids for WASF3 sgRNA were: 5’-AGC TAT ACC TGT ATG GTG TC-3’ and 5’-GAG GCG GTG GCT TAT CAC TC-3’. Plasmids construction was performed as described (20).
Patient samples
One hundred and thirty-four GC patients were included in our study, which were diagnosed and underwent surgery in Peking University Cancer Hospital & Institute between 2002 and 2007. Gastric tumours from each patient were formalin-fixed and paraffin-embedded. Postoperative follow-up was lasted at least 3 years for 134 GC patients. None of the patients received any preoperative treatment. The overall survival was calculated from the date of operation to that of patient death or the latest date of follow-up. In our 134 GC patients, the mean survival time is 36.6 months, and 67.9% (91/134) patients died during the whole follow-up periods.
IHC
GC tissue sections (4 µm) were baked at 80 °C for 2 hours, then dewaxed with xylene and rehydrated with graded alcohol washes. After performing antigen retrieval in a microwave, following to block endogenous peroxidase activity for 10 min with 3% hydrogen peroxide in methanol. Ten percent fetal bovine serum was used to block the non-specific binding for half an hour at 37 °C. The sections were incubated with primary antibody (1:100, ab-110739, Abcam Inc., Cambridge, UK) at 4 °C. For the negative controls, PBS was added instead of primary antibody. Immunostaining was detected with two-step diaminobenzidine visualization (Dako, Glostrup, Denmark). Histopathological sections were examined and scored by two independent pathologists who were blind with the clinical data of patients. The results of IHC were evaluated according to the degree of staining and the percentage of positive cells. According to the degree of staining, each section was defined as three points [1, 2, 3], with “1” light or slightly above background; “2” medium or significantly above background; “3” points for strong staining. According to the percentage of positive tumor cells, each section was classified by four groups [0, 1, 2, 3], with “0” less than 10%; “1” 11–30%; “2” 31–60% and “3” more than 61%.
Final score of each section was calculated by the formula: final score = chromaticity score + positive percentage score. The final score <2 is defined as negative(−); 2–3 as weak positive(+); 4–5 as medium positive(++) and >6 as strong positive(+++). In this study, WASF3 expression was either defined as low-expression group (− and +), or as high-expression group (++ and +++).
Western-blot analysis
GC cells were lysed with RIPA buffer (Pierce Biotechnology, Rockford, IL, USA) with a protease inhibitor cocktail (Roche, Basel, Switzerland). The Western-blot was performed using standard procedures. Gel-separated proteins were transferred to 0.22 µm polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA, USA), then incubated in WASF3 antibody (1:500, Abcam Inc., Cambridge, UK) at 4 °C overnight. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody was used as an internal control. Immunoreactive bands were visualized with a chemiluminescence detection system (Pierce, Rockford, IL, USA).
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA of GC cells was extracted with Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions, and cDNA was synthesized by the AMV cDNA Synthesis kit (Promega, Madison, WI, USA). The mRNA expression of WASF3 was detected by qRT-PCR. The primers of WASF3 were listed as follows: WASF3: 5’-TTC TAG CTC ACT GCT TTC AGG-3’ (sense) and 5’-TGG CCT TCT CCA TTC ATT TT-3’ (antisense). GAPDH: 5’-GAA GGT GAA GGT CGG AGT-3’ (antisense) and 5’-GAA GAT GGT GAT GGG ATT TC-3’ (antisense). The relative expression of WASF3 was standardized with GAPDH expression levels. All reactions were performed in triplicate.
Cell proliferation assay
GC cells (4×103/well) were plated into 96-well pates for cell proliferation. Cell viability was detected using Cell Counting Kit-8 (CCK-8) assay according to the manufacture’s protocol (Dojindo Molecular Technologies, Beijing, China) and quantified at 4 days, respectively.
Boyden chamber assay
For cell migration and invasion assay, 4×104 cells/Transwell Inserts (Corning Incorporated, New York, NY, USA) were used without or with Matrigel (2 mg/mL, Becton Dickinson, San Jose, CA, USA). For the upper chamber, cells were suspended in serum-free medium. The bottom chamber was added with 700 µL complete medium containing 10% FCS. Cells were allowed to migrate for 24 hours or invade for 48 hours, then remove cells of upper surface and the penetrated cells to the under surface were stained with cell stain solution for 20 minutes. Cell numbers were counted in four randomly selected fields per insert (with 200×).
Statistical analysis
Statistical analysis was carried out with SPSS software (version 16.0, SPSS Inc., Chicago, IL, USA). WASF3 expression in GC tissues and their paired adjacent normal mucosa was calculated by paired two-tailed t-tests. Two-tailed chi-square test and Fisher’s exact test was used to analyse the correlation between clinicopathological parameters and WASF3 expression. Survival curves were calculated with Kaplan-Meier method and the P value for survival was assessed by the log-rank test. The effect of different clinical factors on GC patient survival was performed by univariate and multivariate Cox proportional hazards regression models. The statistical analysis among two experimental groups of cells was calculated with Mann-Whitney test. P<0.05 was considered as statistical significance.
Results
WASF3 protein expression was higher in GC tissues than paired adjacent mucosa
For WASF3 protein expression, 134 primary GC samples (including 62 samples with paired adjacent normal mucosa) was analysed with IHC assay. As a result, the expression of WASF3 predominantly localized on cell membrane and in the cytoplasm (Figure 1A), and WASF3 expressed in most of the GC tissues but little in adjacent normal mucosa (Figure 1A). In addition the rate of WASF3 protein high expression in GC was much higher compared with its in adjacent normal mucosa [Chi-square test P<0.0001, 27.4% (17/62) vs. 4.8% (3/62)] (Figure 1B). These results suggested that WASF3 may correlate with tumorigenesis in GC.

High expression of WASF3 associated with poor differentiation and distant metastasis in GC
By IHC staining, the rate of WASF3 high expression was 26.1% (35/134) in total of 134 GC cases. The relationship between WASF3 expression and clinicopathological parameters was shown in Table 1. Among these 134 patients, WASF3 high expression in tumour samples was found to be significantly correlated with tumour poor differentiation and distant metastasis (P=0.038 and 0.032, respectively) (Table 1) (Figure 1C,D). However, there is no statistical correlation between WASF3 protein expression and age, gender, depth of invasion, lymph node metastasis, vascular invasion, and TNM stage (Table 1). Our results suggested that WASF3 may associate with metastatic phenotype in GC cells.
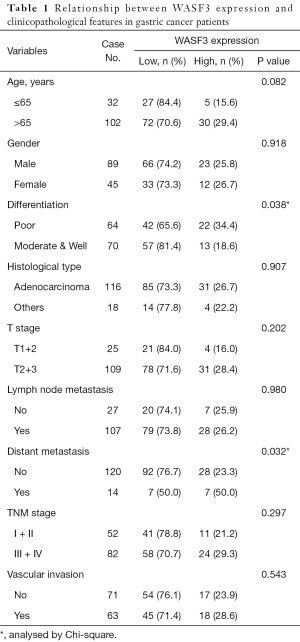
Full table
GC patient with WASF3 high expression predicted poor overall survival
Kaplan-Meier survival analysis demonstrated that patients with WASF3 high expression had a poorer prognosis than those with WASF3 low expression (P=0.044) (Figure 2A). There was the same tendency for the patients prognosis by survival analysis with WASF3 mRNA level (the probe number: 204042_at) from the open database using Kaplan-Meire plotter in GC (http://kmplot.com/analysis/) (Figure 2B). Continually, we tested the clinical parameters to affect the survival of GC patients with and multivariate survival analysis. The univariate results displayed that TNM stage, lymphovascular invasion and WASF3 expression were statistically affected the survival of GC patient, respectively (P<0.001, P=0.034 and 0.047, respectively). Furthermore, the Cox multivariate model indicated that only TNM stage can independently predict GC patient overall survival (P<0.001) (Table 2). These data suggested that WASF3 high expression may predict poor survival of patients with GC.
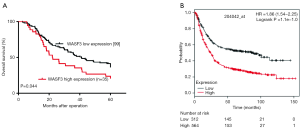
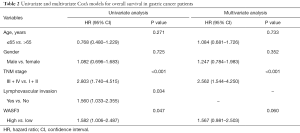
Full table
WASF3 promoted metastatic ability in GC cell in vitro
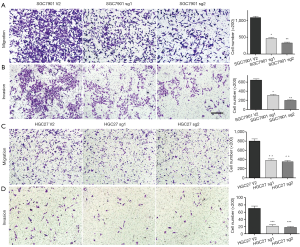
We examined WASF3 expression by Western-blot assay and qRT-PCR in GC cell lines (MKN28, SGC7901, BGC823, and HGC27) and the non-malignant gastric epithelial cell line GES-1, respectively. Our results indicated that WASF3 expression was significantly increased in the most of cancer cells, including SGC7901, BGC823, and HGC27 cells (Figure 3A). To study the role of WASF3 in GC, we chose two specific sgRNAs for WASF3 and transfected to SGC7901 and HGC27 cells to perform the gene knockdown experiments. As a result, the expression of WASF3 was successfully knocked-down by two sgRNAs in both SGC7901 and HGC27 cells using Western-blot assay (Figure 3B). The knockdown of WASF3 did not affect the cell proliferation of SGC7901 and HGC27 cells in vitro (Figure 3C). However, as shown in the Figure 4, the migration and invasion of SGC7901 (Figure 4A,B) and HGC27 (Figure 4C,D) cells were significantly reduced in after knockdown of WASF3 by two different sgRNAs compared to its control cells (CRISPR V2).
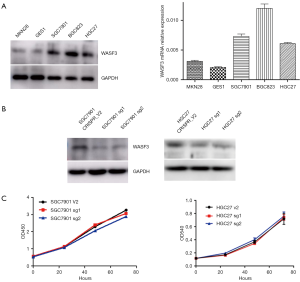
Discussion
Metastasis of cancer plays a key role to result in the majority of cancer deaths. Improvement of surveillance of the metastatic has been developed. Interrupting and prevention of the metastatic process have been dedicated to study to improve the patient outcome. Many metastatic genes have been discovered either as a promoting or suppression function (21,22). This study demonstrates the WASF3 expression status in GC and suggests that WASF3 might play an invasive role in GC. WASF3 is one of the metastatic genes and is reported to promote invasion and metastasis but not in proliferation in breast cancer. Our findings were consistent with these previous reports, demonstrating that WASF3 high expression can change the movement of the cancer cell and enhance their invasive ability both in GC patient clinical data and functional study.
Our results with IHC analysis indicated that WASF3 expression was highly detected in GC tissues compared to their adjacent normal mucosa. Its expression was significantly related with poor differentiation and distant metastasis. But its protein expression pattern was conflict with those in WASF3 mRNA from Gene Expression Omnibus (GEO) database (GSE63089 and GSE13861, respectively, data was not shown) and TCGA database (data was not shown). These might attribute to the post-transcriptional regulation of mRNA. Or the relative normal tissues might have possessed heterogeneity in molecular level other than protein level. Previous studies shown that WASF3 function can be regulated through different mechanisms: transcription suppression, inhibition of activation and reduced stabilization (23-25). Furthermore, patients with high WASF3 expression (in its mRNA and protein level) tended to have a much poorer survival rate, which is consistent with those in breast, but conflict with colorectal cancer (13,17). As we know WASF3 belongs to the WASP family (Wiskott-Aldrich family), which are involved in actin polymerization and lead to cell movement by interrupting actin cytoskeletal dynamics (6). Many molecules involved this process, such as: epidermal growth factor (HER2/HER3), MMP9, RAC, ZEB1, NCKAP1-CYFIP1 and JAK-STAT, etc. (10,26-29). As these molecules could form immunocomplex with WASF3 and perform their different function in various cancer cells. WASF3 might be an intermittent from signal stimulation to cell final movements. The final function of WASF3 may depend on the dominate signal pathway in various cancers.
Furthermore, we continue to calculate WASF3 expression with the survival of GC patient using univariate survival analysis and the Cox multivariate model, respectively. The univariate survival analysis indicated WASF3 expression could affect the survival of GC patients, but unfortunately, it was not an independent factor to predict the GC patient survival. The univariate analysis is just one predictor in the model and the multiple Cox regression has more than one predictor. Cox proportional hazards model, sometimes abbreviated to Cox model, calculates the biological interpretation of the proportional hazards assumption. When the variable is statistic significant in the Cox model, it is an independent risk factor. Thus, it is no wonder that a variable is statistic significant in univariate analysis, but not in the multiple Cox model. In the family of Wiskott-Aldrich, WASF3 just showed its specific function in cell movement. It is an adapter protein to link many other different protein complexes. Both IL6 and HER2 could regulate level of WASF3 transcription through JAK2-STAT signalling pathway (26,29,30). In our present study, WASF3 protein expression was frequently detected in GC cells compared with its matched adjacent noncancerous mucosa. High level of WASF3 expression was more frequently detected in patients with poor differentiation and distant metastasis in GC. It was also one of factors inducing poor survival in GC patients. All these results demonstrated that WASF3 might participate the progression of GC.
In the functional study of WASF3, we found WASF3 involving the biological behaviours through affecting GC cell migration and invasion, but not in proliferation. Knockdown of the WASF3 with CRISPR technology using two different targets significantly suppressed cell movement ability. All these results are consistent with those in breast and prostate cancers. WASF3 might not be a cofactor in cell proliferation, so just regulated itself will not affect the proliferation signals (such as the MEK-ERK axis) (31). Previous reports indicated that WASF3 could downregulated KISS1 transcription level and loss of MMP9 activity through affecting NFkB subunits (p65/p50) (28). Teng et al. recently reports indicated that WASF3 serves as the conduit from HER2/HER3 to signal invasion and metastasis (26). Following stimulation with cytokines or growth factors, WASF3 which interacts with many various molecules will be phosphorylation-activated and proceed its function through actin cytoskeleton in cancer cells (12,29,30,32). These studies suggested WASF3 could be a target for cancer therapy and predict cancer metastasis. Therefore, a larger sample size clinical data in metastasis might be needed for further study.
Till now, there are only two reports for WASF3 in GC (18,19). One reports that miR-218 inhibits cell proliferation, migration and epithelial mesenchymal transformation (EMT) by targeting gene WASF3 (18), and gets the results that restoration of WASF3 expression in the miR-218 over-expression SGC7901 cells impairs miR-218-induced inhibition of proliferation, migration, and EMT in SGC7901 cells. In our study, we perform the knock-down expression of WASF3 in SGC7901 and HGC27 cells, and our data suggest that that knock-down of WASF3 inhibits the movement, but not the proliferation of GC cells. Since the miR-218 over-expression SGC7901 cells are different from the wild type of SGC7901 cells, our result does not conflict with the previous study. The other reports that WASF3 promotes GC cell EMT by increasing Snail molecules (19), and draws the results that over-expression of WASF3 in the SGC7901 cells promotes the cell proliferation and migration. Although, over-expression of WASF3 promotes cell proliferation of SGC7901 cells in their study, we fail to find that knock-down of WASF3 inhibits cell proliferation of SGC7901 cells in our study. In addition to gene expression, there are many other factors affect cell proliferation, including chemokines, hormone and growth factors and cytokine. Thus, it might attribute to the different cell culture condition.
In conclusion, we have demonstrated WASF3 protein expressed highly in cancer tissues and correlated with patient distant metastasis in GC. WASF3 might be a promising metastatic predictor together with some cytokines or growth factors during GC progression. However, these related studies should need us continue to explore. Our findings suggested that WASF3 might be a useful target for future treatment interrupting cancer cell progression.
Acknowledgments
Funding: This study was supported by Beijing Municipal Administration of Hospitals Clinical Medicine Development of special funding support (No. ZYLX201504); Beijing Health System of High Level Health Technical Personnel Training Project (No. 2013-3-067).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.03.13). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study had been approved by the Research and Ethical Committee of Peking University Cancer Hospital & Institute (No. 2017KT79). A written informed consent had been obtained from each patient participated in this study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen W, Zheng R, Zhang S, et al. Cancer incidence and mortality in China in 2013: an analysis based on urbanization level. Chin J Cancer Res 2017;29:1-10. [Crossref] [PubMed]
- Chen W, Zheng R, Zhang S, et al. Cancer incidence and mortality in China, 2013. Cancer Lett 2017;401:63-71. [Crossref] [PubMed]
- Shin A, Kim J, Park S. Gastric cancer epidemiology in Korea. J Gastric Cancer 2011;11:135-40. [Crossref] [PubMed]
- Lin Y, Ueda J, Kikuchi S, et al. Comparative epidemiology of gastric cancer between Japan and China. World J Gastroenterol 2011;17:4421-8. [Crossref] [PubMed]
- Nakajima T, Ota K, Ishihara S, et al. Meta-analysis of 10 postoperative adjuvant chemotherapies for gastric cancer in CIH. Gan To Kagaku Ryoho 1994;21:1800-5. [PubMed]
- Sossey-Alaoui K, Su G, Malaj E, et al. WAVE3, an actin-polymerization gene, is truncated and inactivated as a result of a constitutional t(1;13)(q21;q12) chromosome translocation in a patient with ganglioneuroblastoma. Oncogene 2002;21:5967-74. [Crossref] [PubMed]
- Sossey-Alaoui K, Safina A, Li X, et al. Down-regulation of WAVE3, a metastasis promoter gene, inhibits invasion and metastasis of breast cancer cells. Am J Pathol 2007;170:2112-21. [Crossref] [PubMed]
- Chen Z, Borek D, Padrick SB, et al. Structure and control of the actin regulatory WAVE complex. Nature 2010;468:533-8. [Crossref] [PubMed]
- Chen B, Brinkmann K, Chen Z, et al. The WAVE regulatory complex links diverse receptors to the actin cytoskeleton. Cell 2014;156:195-207. [Crossref] [PubMed]
- Teng Y, Bahassan A, Dong D, et al. Targeting the WASF3-CYFIP1 Complex Using Stapled Peptides Suppresses Cancer Cell Invasion. Cancer Res 2016;76:965-73. [Crossref] [PubMed]
- Teng Y, Qin H, Bahassan A, et al. The WASF3-NCKAP1-CYFIP1 Complex Is Essential for Breast Cancer Metastasis. Cancer Res 2016;76:5133-42. [Crossref] [PubMed]
- Sossey-Alaoui K, Li X, Ranalli TA, et al. WAVE3-mediated cell migration and lamellipodia formation are regulated downstream of phosphatidylinositol 3-kinase. J Biol Chem 2005;280:21748-55. [Crossref] [PubMed]
- Kulkarni S, Augoff K, Rivera L, et al. Increased expression levels of WAVE3 are associated with the progression and metastasis of triple negative breast cancer. PLoS One 2012;7:e42895 [Crossref] [PubMed]
- Wu J, Wang GC, Chen XJ, et al. Expression of WASF3 in patients with non-small cell lung cancer: Correlation with clinicopathological features and prognosis. Oncol Lett 2014;8:1169-74. [Crossref] [PubMed]
- Teng Y, Ren MQ, Cheney R, et al. Inactivation of the WASF3 gene in prostate cancer cells leads to suppression of tumorigenicity and metastases. Br J Cancer 2010;103:1066-75. [Crossref] [PubMed]
- Taylor MA, Davuluri G, Parvani JG, et al. Upregulated WAVE3 expression is essential for TGF-beta-mediated EMT and metastasis of triple-negative breast cancer cells. Breast Cancer Res Treat 2013;142:341-53. [Crossref] [PubMed]
- Zhang Y, Guan XY, Dong B, et al. Expression of MMP-9 and WAVE3 in colorectal cancer and its relationship to clinicopathological features. J Cancer Res Clin Oncol 2012;138:2035-44. [Crossref] [PubMed]
- Yue Z, Feng W, Xiangke L, et al. WAVE3 promotes epithelial-mesenchymal transition of gastric cancer through upregulation of Snail. Cancer Gene Ther 2014;21:499-506. [Crossref] [PubMed]
- Wang G, Fu Y, Liu G, et al. miR-218 Inhibits Proliferation, Migration, and EMT of Gastric Cancer Cells by Targeting WASF3. Oncol Res 2017;25:355-64. [Crossref] [PubMed]
- Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 2014;11:783-4. [Crossref] [PubMed]
- Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell 2011;147:275-92. [Crossref] [PubMed]
- Nguyen DX, Massagué J. Genetic determinants of cancer metastasis. Nat Rev Genet 2007;8:341-52. [Crossref] [PubMed]
- Ghoshal P, Teng Y, Lesoon LA, et al. HIF1A induces expression of the WASF3 metastasis-associated gene under hypoxic conditions. Int J Cancer 2012;131:E905-15. [Crossref] [PubMed]
- Teng Y, Ngoka L, Mei Y, et al. HSP90 and HSP70 proteins are essential for stabilization and activation of WASF3 metastasis-promoting protein. J Biol Chem 2012;287:10051-9. [Crossref] [PubMed]
- Teng Y, Ren X, Li H, et al. Mitochondrial ATAD3A combines with GRP78 to regulate the WASF3 metastasis-promoting protein. Oncogene 2016;35:333-43. [Crossref] [PubMed]
- Teng Y, Pi W, Wang Y, et al. WASF3 provides the conduit to facilitate invasion and metastasis in breast cancer cells through HER2/HER3 signaling. Oncogene 2016;35:4633-40. [Crossref] [PubMed]
- Teng Y, Liu M, Cowell JK. Functional interrelationship between the WASF3 and KISS1 metastasis-associated genes in breast cancer cells. Int J Cancer 2011;129:2825-35. [Crossref] [PubMed]
- Teng Y, Mei Y, Hawthorn L, et al. WASF3 regulates miR-200 inactivation by ZEB1 through suppression of KISS1 leading to increased invasiveness in breast cancer cells. Oncogene 2014;33:203-11. [Crossref] [PubMed]
- Teng Y, Ghoshal P, Ngoka L, et al. Critical role of the WASF3 gene in JAK2/STAT3 regulation of cancer cell motility. Carcinogenesis 2013;34:1994-9. [Crossref] [PubMed]
- Teng Y, Ross JL, Cowell JK. The involvement of JAK-STAT3 in cell motility, invasion, and metastasis. JAKSTAT 2014;3:e28086 [Crossref] [PubMed]
- Geest CR, Buitenhuis M, Groot Koerkamp MJ, et al. Tight control of MEK-ERK activation is essential in regulating proliferation, survival, and cytokine production of CD34+-derived neutrophil progenitors. Blood 2009;114:3402-12. [Crossref] [PubMed]
- Suetsugu S, Miki H, Takenawa T. Identification of two human WAVE/SCAR homologues as general actin regulatory molecules which associate with the Arp2/3 complex. Biochem Biophys Res Commun 1999;260:296-302. [Crossref] [PubMed]


