
Circular RNA FBXW7: implication in glioma tumorigenesis
The significance of circular RNAs (circRNAs) in humans and other higher eukaryotes has gained notable attention in the last decade, with the discovery rate and association of novel circRNAs with specific biological processes continuously increasing. circRNAs were initially regarded as non-functional byproducts or errors of posttranscriptional processing and were considered as a redundant, lowly expressed “splicing noise”. However, these initial underestimations of circRNA quantities and functionality were negated by the advances in the development of global transcriptome analysis and bioinformatics. In particular, RNA deep sequencing technology (RNA-seq) in combination with state-of-the-art detection tools and software packages for complex, large-scale RNA-seq data analysis. Utilization of RNA-seq has enabled a significant breakthrough in identification, characterization, and annotation of novel circRNAs and their association with specific molecular pathways and metabolic processes in humans and other eukaryotes (1-4). Thus, it became clear that circRNAs are abundant and widespread in a variety of organisms and appear to be stably expressed in a cell- and tissue-dependent and, also, developmental stage-specific manner (1,3-6).
circRNAs are classified as a unique subclass of non-coding RNAs (ncRNAs) and are characterized by their covalently closed continuous loop structures, lacking terminal 5’ caps and 3’ polyadenylated tails. They are predominantly generated in the process of spliceosome-mediated back-splicing and can be formed from different genomic regions, including exons, introns and noncoding, antisense, 5’-UTR, 3’-UTR or intergenic genomic regions (1,7,8). Acquisition of evidence regarding circRNA functionality has been the essence of many studies, within which, circRNAs have been mainly recognized as important regulators of gene expression. As such, circRNAs may act as microRNA (miRNA) sponges, sponges for RNA-binding proteins (RBPs), including those that regulate transcription and, also, as regulators of translation (9-11). In addition to their regulatory functions, the circRNA protein-coding ability has been revealed just recently, implying that endogenous circRNAs may represent a novel type of protein-coding RNAs (12-14).
With ever-growing information on human circRNA abundance, differential expression and spatiotemporal presence in cells, tissues and body fluids, dysregulation in circRNA expression profiles may serve as a useful indicator for determining irregularities in human development and for assessing various disease states, including cancer. As demonstrated, dysregulated circRNA expression patterns in various cancerous cells and tissues showed a significant correlation with cancer patient’s clinical characteristics, suggesting they may be utilized to effectively differentiate cancer patients from healthy individuals. Furthermore, several circRNAs have been tested for their diagnostic performance and have been proposed as potential novel biomarkers for cancer diagnosis and prognosis (15). However, there still exists a necessity for an in-depth assessment of functionality for the majority of identified circRNAs, since information regarding their function and protein-coding ability in vivo remains relatively insufficient. Thus, to identify key cancer-associated circRNAs and to develop effective circRNA-associated cancer screening strategies, more functional studies comprising detailed circRNA-miRNA-mRNA-protein interaction network analyses would be essential.
Several studies have revealed a high abundance of circRNAs in neuronal tissues, especially in the synaptic regions. As shown, almost one-fifth of protein-coding genes in the brain produce circRNAs, and it is assumed that these circRNAs are among key regulators of normal neuronal development and function in humans (16). Therefore, altered expression of brain-specific circRNAs may result in diverse neuronal disorders, including brain cancer. Gliomas are among the most common primary malignant brain tumors in adults and are characterized by a relatively high patient’s morbidity and mortality (17). In recent years, there has come to a breakthrough in understanding circRNA involvement in regulating glioma. Indeed, several glioma-associated circRNAs have been characterized to at least some extent, including their expression patterns, miRNA targets and signaling pathways they are involved with (18-21). Intriguingly, a recent study performed by Yang et al. (22), reported the impact of circRNA FBXW7 (circ-FBXW7) and activity of its encoded protein FBXW7-185aa on repressing glioma tumorigenesis. Without a doubt, these findings represent a precedent in revealing the functionality of endogenous circRNA-derived proteins in glioma, carcinogenesis and human biological processes in general.
In the first step of their extensive study, Yang et al. utilized RNA-seq to profile circRNA expression in ten human glioblastomas and their paired adjacent normal tissues. Analysis revealed a total of 31,145 differentially expressed circRNAs between the two groups, all of which were subsequently annotated. They continued by matching the most dysregulated circRNA candidates with the circRNADb database (23) to reveal their potential protein-coding ability and focused their subsequent research on circ-FBXW7, a circularization product of exons 3 and 4 of the FBXW7 gene. By using quantitative real-time PCR (qPCR), downregulation of endogenous circ-FBXW7 expression in glioma samples, when compared to normal controls, was verified and confirmed, showing consistency with their previous RNA-seq analysis. Also, circ-FBXW7 appeared to be predominantly located in the cytoplasm, as most exonic circRNAs (1,3,6). A positive match in circRNADb database and further characterization of circ-FBXW7 protein-coding ability revealed that circ-FBXW7 possessed a spanning junction open reading frame (ORF), encoding a 185-amino acid (approximately 22 kDa) protein, whose translation could be induced by the circ-FBXW7 internal ribosomal entry site (IRES) in a 5'-cap-independent manner. Moreover, FBXW7-185aa shared its amino acid sequence with FBXW7 linear mRNA translated proteins and was capable of inducing cell cycle arrest and inhibited proliferation of glioma cells in vitro and in vivo (22).
Generally, by focusing their research on circ-FBXW7 in glioma, Yang et al. made an important breakthrough in elucidating a cooperative function of mRNA- and circRNA-derived proteins, originating from the same parental gene. As previously determined, the FBXW7 gene encodes the FBXW7α, a well-defined E3 ligase, that suppresses tumorigenesis through targeting c-Myc for ubiquitination-induced degradation (24). In contrast to FBXW7α, FBXW7-185aa does not target c-Myc directly. Instead, FBXW7-185aa competitively interacts with USP28, a deubiquitination enzyme that impairs FBXW7α activity toward c-Myc. Thereby, by antagonizing USP28, FBXW7-185aa enables FBXW7α-mediated ubiquitination of c-Myc, indirectly promoting its degradation (Figure 1) (22). These findings imply that circ-FBXW7 has a pivotal tumor-suppressive role as a novel protein-coding RNA, instead of being a regulatory ncRNA. Also, linear and circular transcripts of the FBXW7 gene with their encoded proteins appear to complement each other in their tumor-suppressive activity (Figure 1). Thus, an important question remains unanswered—are there more circRNAs with similar features present in human tissues, which remained so far unnoticed? Regarding the abundance and widespread of circRNAs, such presumptions are plausible, and we may surely anticipate more comprehensive studies aiming to address this issue in the near future. In time, other translatable circRNAs and their proteins may gradually become revealed and associated with specific biological processes, including carcinogenesis.
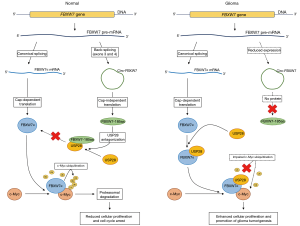
Nevertheless, as emphasized by the authors, small sample size remains the crucial limitation of the study on circ-FBXW7 (22). Yang et al. proposed circ-FBXW7 and FBXW7-185aa as potential independent prognostic biomarkers for glioblastoma. Both, circ-FBXW7 and FBXW7-185aa levels were significantly downregulated in glioblastoma clinical samples, and higher circ-FBXW7 expression positively correlated with glioblastoma patient overall survival time. However, clinical implications of circ-FBXW7 were assessed only in 38 glioblastoma samples and their paired periphery normal brain tissues. Therefore, circ-FBXW7 and FBXW7-185aa diagnostic performance (including sensitivity, specificity and the area under the receiver operating characteristic curve analysis; AUC) should be further validated across a significantly larger patient cohort, before being accepted as reliable prognostic biomarkers for glioma in clinical practice.
To conclude, the study by Yang et al. revealed a thus far unidentified mechanism of circRNA function in human brain cancer and took an important step toward a more comprehensive understanding of the functionality of endogenous circRNAs and their translated proteins. Findings on circ-FBXW7 and FBXW7-185aa in glioma suggest that human circRNAs and circRNA-derived proteins, in combination with proteins originating from linear mRNA isoforms, may have an important complementary role in regulating cellular proliferation and might also be implicated in tumorigenesis. Potential discovery and characterization of circRNAs with similar properties in other cell types and tissues would, therefore, without a doubt, enable significant progress in developing novel, improved cancer treatment strategies.
Acknowledgments
We would like to acknowledge Dr. Michael Dean for proofreading our editorial.
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Chunlin Ou (Cancer Research Institute, Central South University, Changsha, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.04.06). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Wang PL, Bao Y, Yee MC, et al. Circular RNA is expressed across the eukaryotic tree of life. PLoS One 2014;9:e90859 [Crossref] [PubMed]
- Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012;7:e30733 [Crossref] [PubMed]
- Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development 2016;143:1838-47. [Crossref] [PubMed]
- Salzman J, Chen RE, Olsen MN, et al. Cell-type specific features of circular RNA expression. PLoS Genet 2013;9:e1003777 [Crossref] [PubMed]
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013;19:141-57. [Crossref] [PubMed]
- Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife 2015;4:e07540 [Crossref] [PubMed]
- Starke S, Jost I, Rossbach O, et al. Exon circularization requires canonical splice signals. Cell Rep 2015;10:103-11. [Crossref] [PubMed]
- Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015;22:256-64. [Crossref] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. [Crossref] [PubMed]
- Hentze MW, Preiss T. Circular RNAs: splicing’s enigma variations. EMBO J 2013;32:923-5. [Crossref] [PubMed]
- Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRNAs. Mol Cell 2017;66:9-21.e7. [Crossref] [PubMed]
- Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell 2017;66:22-37.e9. [Crossref] [PubMed]
- Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res 2017;27:626-41. [Crossref] [PubMed]
- Bolha L, Ravnik-Glavac M, Glavac D. Circular RNAs: Biogenesis, Function, and a Role as Possible Cancer Biomarkers. Int J Genomics 2017;2017:6218353 [Crossref] [PubMed]
- Chen W, Schuman E. Circular RNAs in Brain and Other Tissues: A Functional Enigma. Trends Neurosci 2016;39:597-604. [Crossref] [PubMed]
- Ostrom QT, Bauchet L, Davis FG, et al. The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol 2014;16:896-913. [Crossref] [PubMed]
- Song X, Zhang N, Han P, et al. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res 2016;44:e87 [Crossref] [PubMed]
- Zheng J, Liu X, Xue Y, et al. TTBK2 circular RNA promotes glioma malignancy by regulating miR-217/HNF1beta/Derlin-1 pathway. J Hematol Oncol 2017;10:52. [Crossref] [PubMed]
- Yang P, Qiu Z, Jiang Y, et al. Silencing of cZNF292 circular RNA suppresses human glioma tube formation via the Wnt/β-catenin signaling pathway. Oncotarget 2016;7:63449-55. [PubMed]
- Li G, Yang H, Han K, et al. A novel circular RNA, hsa_circ_0046701, promotes carcinogenesis by increasing the expression of miR-142-3p target ITGB8 in glioma. Biochem Biophys Res Commun 2018;498:254-61. [Crossref] [PubMed]
- Yang Y, Gao X, Zhang M, et al. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J Natl Cancer Inst 2018;110. [PubMed]
- Chen X, Han P, Zhou T, et al. circRNADb: A comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep 2016;6:34985. [Crossref] [PubMed]
- Yada M, Hatakeyama S, Kamura T, et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J 2004;23:2116-25. [Crossref] [PubMed]


