
Identification of novel genes and pathways in colorectal cancer exosomes: a bioinformatics study
Introduction
Colorectal cancer (CRC) is one of the most frequent malignant tumors with 1.4 million new cases and is the fourth most common cause of oncological death throughout the world (1,2). Despite the incidence and mortality rates decreased over the past several decades, more than one third of CRC patients will develop into metastatic disease (3). Thus, mechanism underlies pathogenesis and metastasis of CRC still requires a further exploration.
Exosomes, which are secreted into the extracellular milieu by various cell types, are phospholipid bilayer nanovesicles ranging from 30 to 100 nm in diameter (4). Tumor-derived exosomes (TDEs) can carry various molecular cargo including proteins, DNA and all types of RNA and then mediate signal transduction in neighboring cells or distant anatomic locations (5). TDEs perform crucial roles in the formation of the “pre-metastatic niches” and initiation of the epithelial-mesenchymal transition (EMT) (6,7). Soldevilla et al. have shown that CRC exosomes enriched in ΔNp73 mRNA can diffuse into recipient cell and promote proliferation (8).
With the development of high-throughput microarray and sequencing, pathway-based methods have been the first choice for complex disease analysis, particularly malign tumors (9). Recently, several studies using CRC tissues and matched normal tissues have found out numerous differentially expressed genes (DEGs) through microarray analysis, which contribute to explaining the underlying molecular mechanism of CRC (10-12). However, the correlation between DEGs of CRC exosomes and disease pathogenesis remains unknown. Thus, we conducted a comprehensive bioinformatics analysis of CRC exosomes microarray studies to discover key DEGs in exosomes related to pathogenesis and metastasis of CRC.
Methods
Data source
The gene expression profiles of GSE100206 and GSE100063 were downloaded from exoRBase (http://www.exoRBase.org) which collects RNA-seq data analysis of human blood exosomes (13,14). Totally 32 normal exosome specimens were enrolled in GSE100206, while 12 CRC exosome specimens were enrolled in GSE100063.
The gene expression profile of GSE32323 was downloaded from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) database (15). This dataset contains 17 pairs of cancer and non-cancerous tissues from CRC patients.
Date processing and screening for DEGs
For the process of GSE100206 and GSE100063, Trimmomatic was used to remove adapters and low-quality bases from the raw sequencing reads. Read counts for each gene were normalized to TPM values (transcript per million). Detailed methods can be found in this paper (14). We used affy package in R to normalize the raw data of GSE32323(16).
The DEGs between CRC group and normal group were estimated using limma package in R statistical software (17). Only genes met the cut-off criteria of P<0.05 and |log2 fold change (FC)| ≥1.0 were regarded as DEGs.
Enrichment analysis of DEGs
DAVID (The Database for Annotation, Visualization and Integrated Discovery, https://david.ncifcrf.gov/), as a program used for genes or proteins functional annotation, was adopted for functional and pathway enrichment analysis (18). Based on selected DEGs, we performed gene ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis with the thresholds of P<0.05 (19,20).
Protein-protein interaction (PPI) network construction
We used Search Tool for the Retrieval of Interacting Genes (STRING) database to recognize potential interactions among DEGs with a combined score >0.4 (21). Then the PPI network was visualized by Cytoscape software (22). To find dense clique-like structures within a network, Molecular Complex Detection (MCODE) was conducted with MCODE score >3 and number of nodes >4. The hub proteins were identified on the basis of degree and betweenness value using cytoHubba (23,24).
Functional annotation of DEGs
Catalogue Of Somatic Mutations In Cancer (COSMIC, http://cancer.sanger.ac.uk/census) database provides detailed annotation for each tumor-associated gene (TAG) including oncogenes and tumor suppressor genes (TSGs) (25). Functional annotation of DEGs was performed to detect oncogenes and TSGs among DEGs. Up-regulated oncogenes and down-regulated TSGs were regarded as positive genes for tumor formation, otherwise, TAGs were defined as negative genes.
Overall survival (OS) analysis
LinkedOmics (http://www.linkedomics.org) is a publicly available portal that includes multi-omics data from all 32 TCGA cancer types (26). We used cox regression analysis to evaluate the relationship between selected DEGs expression and CRC patients’ OS. Then, the CRC patients were divided into high- and low-expression groups according to the median mRNAs expression level. Log-rank P value was calculated to compare the survival distribution between two groups.
Statement of ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors. Ethics approval is not required for this study.
Statistical analysis
Chi-square test was used to determine the statistical significance of TAGs number. All statistical analysis was performed using STATA12 (StataCorp, College Station, TX, USA), and a P value of <0.05 was considered to be statistically significant.
Results
Identification of DEGs
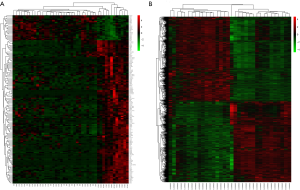
A total of 149 DEGs (127 up-regulated and 22 down-regulated) were obtained in the tumor exosomes group according to the cut-off value for screening. We also identified 1,507 DEGs from the tumor tissues group, which included 771 up-regulated and 736 down-regulated DEGs. The representative heatmap of DEGs in tumor exosomes and tissues were illustrated in Figure 1A,B respectively.
GO and pathway enrichment analysis
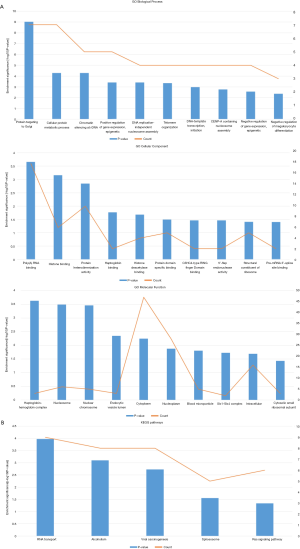
To annotate the biological function of the DEGs in the tumor exosomes, we performed GO and KEGG pathway analysis. The top 10 terms of DEGs were displayed in Figure S1A. GO analysis revealed that the DEGs were significantly involved in biological processes (BP) of response to protein targeting, protein metabolism and gene expression. As for molecular function (MF), enrichment indicate protein and histone binding, endonuclease activity and pre-mRNA splice site binding. Besides, cell component (CC) enrichment displayed endocytic vesicle lumen, cytoplasm and blood microparticle, which hinted that DEGs might play a critical role in exosomes formation.
KEGG pathway analysis suggested that the DEGs were enriched in 5 pathways including RNA transport, alcoholism, viral carcinogenesis, spliceosome and Ras signaling pathway (Figure S1B).
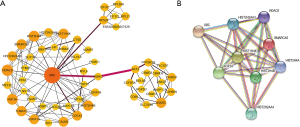
PPI network construction
A total of 116 PPI relationships were constructed on the basis of STRING database (Figure 2A). In the network, 5 node proteins, including UBC, H3F3A, HIST2H2AA3, AKT3, and HSPA1B, which showed a strong association with other node proteins, were selected as hub proteins. As shown in Figure 2B, one significant module was selected with the criteria of number of nodes >4. The key module showed functions enriched in pathways such as alcoholism and viral carcinogenesis (Table 1).

Full table
Functional annotation of DEGs
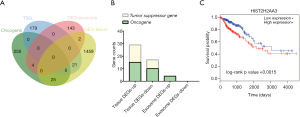
As shown in Figure 3A, 21 TSGs and 25 oncogenes were found in tumor tissues with 4 oncogenes (H3F3A, U2AF1, P2RY8 and APOBEC3B) in tumor exosomes. However, there was no significant difference in the ratio of TSGs to oncogenes among DEGs of tumor tissues or exosomes (P=0.21). Subgroup analysis was conducted in up-regulated and down-regulated DEGs (Figure 3B, Table 2). According to our definition of positive genes (up-regulated oncogenes and down-regulated TSGs) and negative genes (up-regulated TSGs and down-regulated oncogenes), we found 22 positive genes and 24 negative genes in tumor tissues with 4 positive genes in tumor exosomes. We did not find any significant difference either (P=0.14). Subsequently, OS analysis showed that higher HIST2H2AA3 expression level was significantly associated with lower OS (P=0.0015, Figure 3C).

Full table
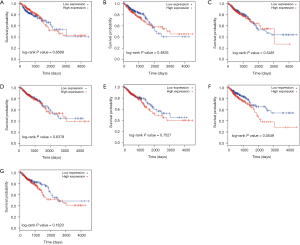
However, no significant differences in OS for H3F3A, U2AF1, P2RY8, APOBEC3B, UBC, AKT3, and HSPA1B were observed (Figure S2).
Discussion
Identifying abnormally expressed genes associated with CRC would greatly benefit the diagnosis and therapies of this disease. In this study, a total of 149 DEGs were identified in CRC exosomes relative to normal exosomes through analyzing gene expression profile of GSE100206 and GSE100063. Enrichment analysis of DEGs might provide novel insights for unraveling mechanism of CRC development and progression.
As was suggested by DAVID analysis, the significant GO terms were related to positive regulation of gene expression, protein binding to Golgi, cellular protein metabolic process and endonuclease activity. It is reasonable that sustaining proliferative signaling and deregulating cellular energetics are hallmarks of cancers including CRC (27). Regard to the signaling pathway, we found that RNA transport, alcoholism, viral carcinogenesis, spliceosome and Ras signaling pathway were highly enriched. It is consistent with the fact that carcinogens induce long-lasting increased in insoluble NTPase may alter the RNA transport associated with cancer and carcinogenesis (28,29). Dysregulation of spliceosome gene expression including U2AF1 is also associated with tumor formation and cell survival (30). Viral pathogens and alcoholism have also been found to increase risk of CRC development (31,32). Ras signaling pathway enriched in AKT3 is activated in many malignancies and has become a major focus of drug targeting efforts (33).
In PPI network analysis, top 5 hub genes appeared to be UBC, H3F3A, HIST2H2AA3, AKT3, and HSPA1B. As the most significant hub gene, the detailed function of UBC for CRC has yet been reported to data. Due to the lowest levels of expression variability and constitution in CRC and other tissues, UBC, a ubiquitin gene, is used as the housekeeping genes (34). When compared the expression of UBC between CRC tissues and matched normal tissues, there was no significant difference at protein level (35). However, ubiquitination has been associated with CRC (36). Thus, further molecular research may focus on the role of UBC in exosomes instead of tumor tissues. H3F3A and HIST2H2AA3 are histone genes belonging to the H3A and H2A family respectively. They constitute the octamer of core histone proteins with H2B, H3 and H4. Histones are basic nuclear proteins that are responsible for the nucleosome structure (37). H3F3A mutations often occur in gliomas especially high-grade gliomas and reprogram epigenetic landscape (38). Decreased expression of HIST2H2AA3 at the protein and mRNA levels has been reported in the hepatocellular carcinoma (39). AKT3 gene encodes a protein belonging to Akt kinase family, which exert functions in cell proliferation, differentiation, apoptosis, tumorigenesis, as well as glycogen synthesis and glucose uptake. Compared with ubiquitous expression Akt1 and Akt2, AKT3 has a more limited tissue distribution. Overexpression of AKT3 occurs in breast cancer and prostate cancer, suggesting that Akt3 is strongly oncogenic (40). Although the role of AKT2 and AKT1 in CRC has been well elucidated, limited information of AKT3 in CRC can be found (41,42). Heat shock protein encoded by HSPA1B is a member of the heat shock protein 70 family. Elevated expression of HSPA1B promotes breast cancer cell growth by arresting cancer cells in G1 (43). A recent meta-analysis suggests HSPA1B ± 1267A/G polymorphism increases risk of hepatocellular carcinoma (44). The roles of all above genes in CRC require further explorations to gain an insight into the function of CRC exosomes.
When we compared the DEGs in tumor exosomes with DEGs in tumor tissues, it was surprised to find similar ratios of TSGs to oncogenes existed in exosomes and tissues. Previous have proven that exosomes mediate the metastasis of cancer (7). Thus, tumor exosomes were expected to contain more oncogenes instead of TSGs. Although we did not find any TSGs in the CRC exosomes, the research conducted by Teng et al. may offer a novel explanation of TSGs enriched in exosomes if they do exist in exosomes (45). Selective sorting of TSGs into exosomes with more oncogenes left in tumor cells can also promote primary tumor progression. Moreover, it should be remembered that exosomes used in GSE100206 and GSE100063 were not tumor-specific exosomes. Therefore, the results can be affected by the mixed exosomes in blood (46).
Although some key genes and pathways were identified, several limitations should be acknowledged in this study. The clinical prognosis analyzed in our study used the date from tumor tissues but not the exosomes due to the availability of data. Besides, we could not define the biological function of tumor derived exosome without a cancer exosome-specific biomarker. Further molecular experiments should be conducted to better confirm our findings of the important genes and pathways in CRC.
In conclusion, our study revealed a number of DEGs in the CRC exosomes by bioinformatics analysis. The data from this study may contribute to future translational medicine studies in identifying new approaches for the diagnosis and treatment of CRC.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.05.32). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This article does not contain any studies with human participants or animals performed by any of the authors. Ethics approval is not required for this study and informed consent was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Xu P, Zhu Y, Sun B, et al. Colorectal cancer characterization and therapeutic target prediction based on microRNA expression profile. Sci Rep 2016;6:20616. [Crossref] [PubMed]
- Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin 2016;66:271-89. [Crossref] [PubMed]
- van den Boorn JG, Dassler J, Coch C, et al. Exosomes as nucleic acid nanocarriers. Adv Drug Deliv Rev 2013;65:331-5. [Crossref] [PubMed]
- Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev 2013;32:623-42. [Crossref] [PubMed]
- Costa-Silva B, Aiello NM, Ocean AJ, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol 2015;17:816-26. [Crossref] [PubMed]
- Syn N, Wang L, Sethi G, et al. Exosome-Mediated Metastasis: From Epithelial-Mesenchymal Transition to Escape from Immunosurveillance. Trends Pharmacol Sci 2016;37:606-17. [Crossref] [PubMed]
- Soldevilla B, Rodriguez M, San Millan C, et al. Tumor-derived exosomes are enriched in DeltaNp73, which promotes oncogenic potential in acceptor cells and correlates with patient survival. Hum Mol Genet 2014;23:467-78. [Crossref] [PubMed]
- Surrey LF, Luo M, Chang F, et al. The Genomic Era of Clinical Oncology: Integrated Genomic Analysis for Precision Cancer Care. Cytogenet Genome Res 2016;150:162-75. [Crossref] [PubMed]
- Liu J, Li H, Sun L, et al. Aberrantly methylated-differentially expressed genes and pathways in colorectal cancer. Cancer Cell Int 2017;17:75. [Crossref] [PubMed]
- Sreekumar A, Nyati MK, Varambally S, et al. Profiling of cancer cells using protein microarrays: discovery of novel radiation-regulated proteins. Cancer Res 2001;61:7585-93. [PubMed]
- Carmona FJ, Azuara D, Berenguer-Llergo A, et al. DNA methylation biomarkers for noninvasive diagnosis of colorectal cancer. Cancer Prev Res (Phila) 2013;6:656-65. [Crossref] [PubMed]
- Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 2015;25:981-4. [Crossref] [PubMed]
- Li S, Li Y, Chen B, et al. exoRBase: a database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res 2018;46:D106-12. [Crossref]
- Khamas A, Ishikawa T, Shimokawa K, et al. Screening for epigenetically masked genes in colorectal cancer Using 5-Aza-2'-deoxycytidine, microarray and gene expression profile. Cancer Genomics Proteomics 2012;9:67-75. [PubMed]
- Gautier L, Cope L, Bolstad BM, et al. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004;20:307-15. [Crossref] [PubMed]
- Diboun I, Wernisch L, Orengo CA, et al. Microarray analysis after RNA amplification can detect pronounced differences in gene expression using limma. BMC Genomics 2006;7:252. [Crossref] [PubMed]
- Dennis G Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003;4:3. [Crossref] [PubMed]
- Gene Ontology Consortium. The Gene Ontology (GO) project in 2006. Nucleic Acids Res 2006;34:D322-6. [Crossref] [PubMed]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27-30. [Crossref] [PubMed]
- von Mering C, Huynen M, Jaeggi D, et al. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res 2003;31:258-61. [Crossref] [PubMed]
- Smoot ME, Ono K, Ruscheinski J, et al. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 2011;27:431-2. [Crossref] [PubMed]
- He X, Zhang J. Why do hubs tend to be essential in protein networks? PLoS Genet 2006;2:e88 [Crossref] [PubMed]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 2003;4:2. [Crossref] [PubMed]
- Futreal PA, Coin L, Marshall M, et al. A census of human cancer genes. Nat Rev Cancer 2004;4:177-83. [Crossref] [PubMed]
- Vasaikar SV, Straub P, Wang J, et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 2018;46:D956-63. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Clawson GA, Feldherr CM, Smuckler EA. Nucleocytoplasmic RNA transport. Mol Cell Biochem 1985;67:87-99. [Crossref] [PubMed]
- Wang M, Xiao X, Zeng F, et al. Common and differentially expressed long noncoding RNAs for the characterization of high and low grade bladder cancer. Gene 2016;592:78-85. [Crossref] [PubMed]
- Fei DL, Motowski H, Chatrikhi R, et al. Wild-Type U2AF1 Antagonizes the Splicing Program Characteristic of U2AF1-Mutant Tumors and Is Required for Cell Survival. PLoS Genet 2016;12:e1006384 [Crossref] [PubMed]
- Collins D, Hogan AM, Winter DC. Microbial and viral pathogens in colorectal cancer. Lancet Oncol 2011;12:504-12. [Crossref] [PubMed]
- Phipps AI, Baron J, Newcomb PA. Prediagnostic smoking history, alcohol consumption, and colorectal cancer survival: the Seattle Colon Cancer Family Registry. Cancer 2011;117:4948-57. [Crossref] [PubMed]
- Agarwal E, Brattain MG, Chowdhury S. Cell survival and metastasis regulation by Akt signaling in colorectal cancer. Cell Signal 2013;25:1711-9. [Crossref] [PubMed]
- Watanabe T, Kobunai T, Sakamoto E, et al. Gene expression signature for recurrence in stage III colorectal cancers. Cancer 2009;115:283-92. [Crossref] [PubMed]
- Uhlen M, Zhang C, Lee S, et al. A pathology atlas of the human cancer transcriptome. Science 2017;357: [Crossref] [PubMed]
- Hao Y, Samuels Y, Li Q, et al. Oncogenic PIK3CA mutations reprogram glutamine metabolism in colorectal cancer. Nat Commun 2016;7:11971. [Crossref] [PubMed]
- Monteiro FL, Baptista T, Amado F, et al. Expression and functionality of histone H2A variants in cancer. Oncotarget 2014;5:3428-43. [Crossref] [PubMed]
- Nguyen AT, Colin C, Nanni-Metellus I, et al. Evidence for BRAF V600E and H3F3A K27M double mutations in paediatric glial and glioneuronal tumours. Neuropathol Appl Neurobiol 2015;41:403-8. [Crossref] [PubMed]
- Khare SP, Sharma A, Deodhar KK, et al. Overexpression of histone variant H2A.1 and cellular transformation are related in N-nitrosodiethylamine-induced sequential hepatocarcinogenesis. Exp Biol Med (Maywood) 2011;236:30-5. [Crossref] [PubMed]
- Testa JR, Bellacosa A. AKT plays a central role in tumorigenesis. Proc Natl Acad Sci U S A 2001;98:10983-5. [Crossref] [PubMed]
- Rychahou PG, Kang J, Gulhati P, et al. Akt2 overexpression plays a critical role in the establishment of colorectal cancer metastasis. Proc Natl Acad Sci U S A 2008;105:20315-20. [Crossref] [PubMed]
- Johnson SM, Gulhati P, Rampy BA, et al. Novel expression patterns of PI3K/Akt/mTOR signaling pathway components in colorectal cancer. J Am Coll Surg 2010;210:767-76, 776-8.
- Rohde M, Daugaard M, Jensen MH, et al. Members of the heat-shock protein 70 family promote cancer cell growth by distinct mechanisms. Genes Dev 2005;19:570-82. [Crossref] [PubMed]
- Kuang D, Chen W, Song YZ, et al. Association between the HSPA1B +/-1267A/G polymorphism and cancer risk: a meta-analysis of 14 case-control studies. Asian Pac J Cancer Prev 2014;15:6855-61. [Crossref] [PubMed]
- Teng Y, Ren Y, Hu X, et al. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nat Commun 2017;8:14448. [Crossref] [PubMed]
- Tkach M, Thery C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 2016;164:1226-32. [Crossref] [PubMed]


