
Matrix metalloproteinase (MMP) and immunosuppressive biomarker profiles of seven head and neck squamous cell carcinoma (HNSCC) cell lines
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common form of cancer by incidence worldwide (1). In HNSCC, matrix metalloproteinases (MMPs) degrade extracellular matrix and the basement membrane, which is associated with tumor invasion and metastasis (2). MMPs have also been implicated in tumor progression due to their ability to activate growth factors and enhance angiogenesis (3). Despite this known association, the expression of these MMPs in relation to other biomarkers has not been fully investigated in HNSCC (4).
In HNSCC and other cancers, tumor cells create an immunosuppressive environment through various immunosuppressive mediators and strategies (5-7). Cancer cells use a variety of mechanisms to evade host defenses and metastasize. In this study, we have focused on the tumor microenvironment by examining differences in the biomarkers expressed by cancer cells to induce immunosuppression and the MMPs expressed by cancer cells that aid in metastasis. Specifically, we examined the production of immunosuppressive biomarkers: interleukin-6 (IL-6), vascular endothelial growth factor A (VEGFA), interleukin (IL)-1 alpha (IL-1α), tumor necrosis factor alpha (TNF-α), granulocyte-macrophage colony-stimulating factor (GM-CSF), IL-1 receptor antagonist (IL-1RA), and IL-8; and MMPs: MMP1, MMP7, and MMP9. We also looked at the cell surface marker production of programmed death ligand-1 (PD-L1), Fas ligand (FASL), indoleamine 2,3 dioxygenase (IDO), and cluster of differentiation (CD47). Not only have these biomarkers been suggested as helping tumor cells in creating an immunosuppressive environment, they also contribute to other biological functions like migration, angiogenesis, and cell growth regulation (5).
MMPs are the major enzymes implicated in degradation of the extracellular matrix and the basement membrane (2). MMPs are from a large family of calcium-dependent, zinc-containing endopeptidases. There are 24 MMPs in humans, which are categorized into six groups based on their substrate preferences and domain structure: collagenases (MMP1, 8, 13), gelatinases (MMP2, 9), stromelysins (MMP3, 10), matrilysins (MMP7, 26), membrane-type metalloproteinases (MT-MMPs), and others (2,8). MMPs play important roles in wound healing, tissue repair, angiogenesis, bone remodeling, morphogenesis, tooth eruption, cell communication, and remodeling after injury, but are also implicated in inflammatory diseases such as rheumatoid arthritis, atherosclerosis, and periodontal disease (2).
MMPs also play critical roles in immune responses by either modifying proteins post-translationally to promote rapid delivery to other cells or by inactivating these proteins to initiate or terminate the immune process (9). MMPs can release cytokines, growth factors, and chemokines from their pro-forms (10). These functions allow MMPs to direct systemic inflammation and regulate cytokine biosynthesis through activation of signal transduction pathways. In cancer, MMPs play a role in tumor progression by breaking down the basement membrane barrier leading to metastasis (11). For example, MMP9 is expressed in carcinoma and inflammatory cells in oral squamous cell carcinoma (OSCC, a subtype of HNSCC) tumors (12,13). MMP9 degrades type IV collagen, a main component of the basement membrane, hence its association with cancer metastasis (2,10).
MMP activity is tightly regulated transcriptionally and post-transcriptionally (8). They are also regulated through compartmentalization of MMP release, enzyme activation by removal of the pro-domain, and inhibition by tissue inhibitors of MMPs (TIMPs) or by non-specific proteinase inhibitors. This tight regulation is necessary due to the wide array of substrates. When this regulation is not controlled, it leads to diseases such as cancer, which is why MMPs could serve as potential therapeutic targets (8,14).
The purpose of this study was to elucidate differences in the expression of proteins expressed by HNSCC cancer cell lines, which are representative of differences that could be seen between patient tumor cells, and to better understand the role of biomarkers and MMPs expressed by cancer cells in the tumor microenvironment. Studying these cell responses may lead to potential biomarkers to target for future clinical applications.
Methods
Cell lines
HNSCC cell lines SCC4 (ATCC, Manassas, VA, USA), SCC15 (ATCC), SCC19 (University of Michigan), SCC25 (ATCC), SCC84 (University of Michigan), SCC92 (University of Michigan), and SCC99 (University of Michigan) were used in this study. SCC4, SCC15, SCC25, SCC84, and SCC92 are from the oral cavity, while SCC19 and SCC99 are from the oropharynx (Table 1) (15). TNM classifications of malignant tumors (TNM, where T describes the size of the primary tumor, N describes the nearby lymph nodes, and M describes the metastasis) are listed for the selected cell lines in Table 1. SCC19, SCC84, SCC92, and SCC99 from the University of Michigan have been previously genotyped (16,17). These cell lines were each grown and maintained in a humidified atmosphere of 5% CO2 at 37 °C in T75 flasks. SCC4 cells were grown in complete Dulbecco’s Modified Eagle’s Medium: F-12 (DMEM:F-12) containing 2 mM L-glutamine, 1% nonessential amino acids (ATCC), 400ng/mL hydrocortisone (Sigma-Aldrich Corp., St. Louis, MO, USA), 100 units/mL penicillin (Life Technologies, Madison, WI, USA), 100 units/mL streptomycin (Life Technologies), and 10% fetal bovine serum (ATCC) (18). SCC15, SCC25, and SCC84 cells were grown in complete Lymphocyte Growth Media-3 (LGM-3) (Lonza, Walkersville, MD, USA), 100 units/mL penicillin (Life Technologies), 100 units/mL streptomycin (Life Technologies), and 10% fetal bovine serum (ATCC) (18). SCC19, SCC92, and SCC99 were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 2 mM L-glutamine, 1% nonessential amino acids (ATCC), 100 units/mL penicillin (Life Technologies), 100 units/mL streptomycin (Life Technologies), and 10% fetal bovine serum (ATCC). These media were chosen because they ensured growth and survival of the HNSCC cells.
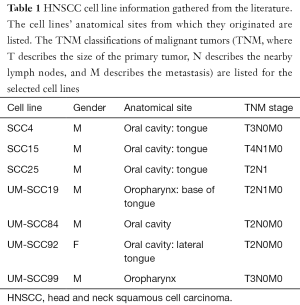
Full table
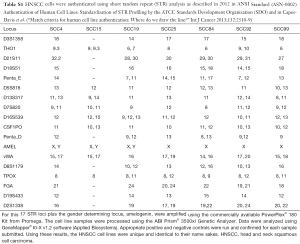
The seven cell lines were authenticated as described in 2012 in ANSI Standard (ASN-0002) Authentication of Human Cell Lines: Standardization of STR Profiling by the ATCC Standards Development Organization (Table S1).
Cell culture supernatants
Once the cells reached approximately 60–80% confluence in T75 flasks, we harvested the adherent HNSCC cells with 0.25% trypsin containing 0.53 mM EDTA (Cat. No. 30-2101, ATCC, Manassas, VA, USA). We counted the cells and tested them for viability using propidium iodide. We centrifuged the cells (400 RCF, Eppendorf 5810R, Westbury, NY, USA) at 4 °C for 10 minutes and resuspended the cells in high-glucose RPMI-1640 with L-glutamine and HEPES (ATCC, Manassas, VA) to 1×106 viable cells/mL. By using this minimal media to characterize the biomarker production for all seven cell lines we can eliminate changes caused by the presence of additives like serum. We added 1.0 mL of this suspension to 3 wells on a 12-well tissue culture plate (Corning Inc., Corning, NY, USA). The plate was incubated in a humidified atmosphere of 5% CO2 at 37 °C. After 24 hours, the cells were examined via tissue culture scope to confirm they had adhered. We removed the tissue culture medium and centrifuged it (3000 RCF, Eppendorf 5415D, Westbury, NY, USA) at room temperature for 5 minutes to remove any residual cells or cell fragments. The supernatant was stored at −80 °C until it was time to determine the concentration of the biomarkers.
Cell lysates
After the supernatant was removed from the wells, we added 1.0 mL of cell lysis buffer (Cell Signaling Technologies, Danvers, MA, USA) containing phenylmethanesulfonyl fluoride (1.0 mM PMSF, Cell Signaling Technologies) to the adhered cell layer in each well. The lysed cell suspension (i.e., cell lysate) was removed and stored at −80 °C.
Biomarker determination
To determine the concentrations of MMP1, MMP7, MMP9, IL-6, VEGFA, IL-1α, TNF-α, GM-CSF, IL-1RA, and IL-8 in cell culture supernatants, we used multiplex immunoassays (R&D Systems, Minneapolis, MN, USA) read on a Luminex100 (Luminex, Madison, WI, USA), which are routinely used in our laboratory (19,20). These immunoassay kits use antibody-coated magnetic beads to bind the desired analytes in a solution and uses a standard curve of known concentrations to determine the unknown concentrations of the samples as previously described (21). We used enzyme-linked immunosorbent assay (ELISA, Cusabio Biotech Co., Ltd., Wilmington, DE, USA) to determine the concentrations of CD47, IDO1, FASL, and PDL1 in cell lysates.
Human papillomavirus (HPV) and p16Ink4a determination
Presence of HPV antigen in the cell lines was assessed using a double-sandwich ELISA (MyBioSource, Inc., San Diego, CA, USA). Both cell lysates and supernatants were tested in the ELISA. Immunohistochemistry (IHC) was used to check p16Ink4a status among the seven cell lines. After the 24-hour incubation in a replicate experiment as the one described above, the supernatant was removed and the remaining adhering cells were fixed in 10% neutral buffered formalin. The cells were then placed in agar and fixed in a paraffin block. Sections of this block were deparaffinized for IHC staining with an antibody for p16Ink4a. Squamous cell carcinoma of the uterus was used as a positive control.
Statistical analysis
Three replications were done for each group and a log-10 transformation was applied to all of the biomarker concentrations to attenuate for positive skew and to make the normality assumption more defensible (19). We first analyzed the log values of the biomarker concentrations using a one-way multivariate analysis of variance (MANOVA) to detect the overall effect for the different cell lines. As the MANOVA reached significance, a univariate one-way ANOVA was performed to explore the impact of the differing cell lines on the concentration within each biomarker, followed by the pairwise group comparisons using the post-hoc Tukey’s honest significant differences test. A 0.05 level was used determine statistically significance.
Results
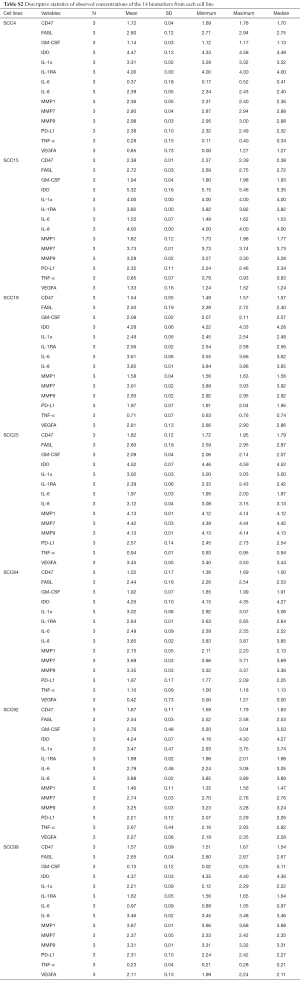
In both the cell supernatants and cell lysates, statistically significant differences were seen among the seven cell lines in all 14 biomarkers (Tables 2,S2). The MANOVA revealed a significant multivariate effect for the different cell lines, Wilk’s Lambda =0.0001 and P<0.0001. The one-way ANOVAs revealed significant differences between the seven cell lines in each of the 14 biomarkers (P<0.05 in each instance).
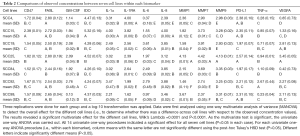
Full table
SCC25 was the highest producer of MMP1, MMP7, and MMP9 (Figure 1, P<0.05). MMP1 production was significantly different in all seven cell lines except SCC19 and SCC92. MMP7 production was significantly different in all seven cell lines except SCC15 and SCC84. MMP9 production was significantly different between several cell lines but not all, with SCC25 being the highest and SCC4 and SCC19 being the lowest. Other than SCC25 being the highest producer of all three MMPs, the only apparent trend is that these cell lines are able to produce more than one type of MMP and at significantly different levels than each other.

GM-CSF and TNF-α, both inflammatory biomarkers, were produced by SCC92 at significantly higher levels than all of the other cell lines, while SCC4 and SCC99 produced the lowest amounts of GM-CSF and TNF-α (Figure 2). SCC15 produced significantly higher amounts of IL-1α and IL-8 than the other six cell lines. IL-1α was produced in moderate amounts by SCC4, SCC25, SCC84, and SCC92 and at significantly lower amounts by SCC19 and SCC99. IL-8 was produced at moderately high amounts in SCC19, SCC84, and SCC92, and SCC4 produced the lowest amount of IL-8. IL-1RA production was highest in SCC4 and SCC15 and lowest in SCC99 and SCC92. IL-6 production was highest in SCC19 and lowest in SCC99 and SCC4. VEGFA production was highest in SCC25 and SCC19 and lowest in SCC4, SCC15, and SCC84. MIP1α, MIP1β, and IL-12 p40 concentrations were also determined, but they were produced at very low levels (Figure S1).
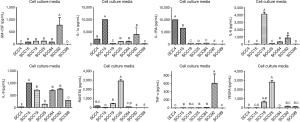
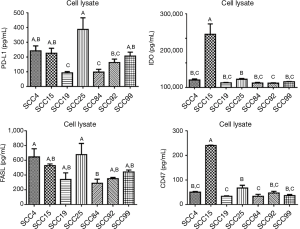
When looking at the cell surface markers detected in the cell lysates, we see that PDL1 production was highest in SCC25 and lowest in SCC19 and SCC84 (Figure 3, P<0.05). This trend was also seen with FASL production, although not significant. SCC15 produced significantly more IDO and CD47 than any of the other cell lines. It is interesting to note that SCC25 was highest in MMP1, MMP7, and MMP9 compared to the other six cell lines (P<0.05) and highest in PDL1 and VEGFA production and SCC15 was highest in IDO, CD47, IL-1α, and IL-8 compared to the other six cell lines (P<0.05).
Human papillomavirus is an emerging risk factor for HNSCC (22). The HPV ELISA showed that these cell lines are HPV negative. Similar studies have also confirmed that four of these cell lines (e.g., SCC4, SCC15, SCC25, and SCC19) were HPV negative (23). The IHC staining revealed that all seven cell lines were negative for p16Ink4a expression, a tumor suppressor protein often used as a surrogate marker for HPV infection (24).
Discussion
As shown here, and in other studies, tumor cells can express several MMPs and at varying amounts (3). Besides contributing to the metastasis of cancer cells, MMPs can contribute to angiogenesis as well; specifically, MMP7 and MMP9 have been shown to have distinct roles in vascularization events taking place in the same tumor. MMP9 can also increase VEGF bioavailability, thereby increasing the amount of VEGF in the tumor microenvironment. MMPs also regulate inflammation in cancer (3). Several MMPs, including MMP1 and MMP9, can cleave pro-TNF-α to activate this proinflammatory cytokine. As we’ve seen here, HNSCC cells can release varying amount of TNF-α. Tumor cells produce TNF-α to promote cell survival. MMP9 can also interact with IL-8 to increase the recruitment of neutrophils to the tumor microenvironment (25).
MMPs also present as desirable drug targets because of the major role they play in disease progression. Early clinical trials for MMP inhibitors have been unsuccessful, but more studies are being conducted using novel approaches to target MMPs in cancer (14). MMPs have also been investigated as prognostic markers for HNSCC. For example, transcription of MMP9 has been proposed as a prognostic marker for treatment response to radiotherapy and chemo-radiotherapy (26). MMP9 has also been proposed as a possible diagnostic marker for early detection of HNSCC (27).
Cytokines, chemokines, and MMPs play a major role in both the inflammatory and tumor microenvironments. These proteins can act on each other and in synchronization. Secretion of transforming growth factor beta (TGF-β), IL-6, IL-10, and TNF-α from OSCC cells has recently been reported to create a favorable environment for tumor growth (28,29). TNF-α has been found to be secreted by stage IV, metastatic, and reoccurrence-derived HNSCC cell lines, suggesting it may serve as an indicator of late stage cancer (29). IL-6 is involved in inflammatory processes indicative of tumor proliferation, but it also has immunosuppressive effects on dendritic cells by preventing dendritic cell maturation (29,30). High expression of IL-6 in OSCC cells has been suggested as a predictive factor of poor response to chemoradiotherapy (31). The anti-inflammatory cytokine IL-1RA, which competes with IL-1β by binding to the IL-1 receptor, is also found in elevated levels in the saliva of patients with OSCC (32). IL-1β is secreted by tumor cells and is a substrate of MMP9 (10). TGF-β and TNF-α are also substrates of MMP9. Programmed death ligand 1 (PDL1) is expressed on tumor cells and its expression can be induced by IFN-γ (33). PDL1 of tumor cells binds to the immune-inhibitory receptor programmed death-1 (PD1) on activated B-cells or T-cells, which allows the tumor cells to evade destruction (33). Vascular endothelial growth factor (VEGF), known to stimulate angiogenesis, is a marker for tumor invasion and metastasis as it too can be detected in HNSCC (29). VEGF may also promote immune tolerance (34). Higher levels of IL-6 and VEGF are produced in late-stage HNSCC cell lines compared to early-stage and metastatic cell lines compared to nonmetastatic cell lines (29).
It was important for us to determine HPV status of our cell lines as HPV is considered a positive risk factor for oropharyngeal squamous cell carcinoma, a subset of HNSCC, and is a current focus of many HNSCC studies (35). While there has been a decrease in the number of tobacco and alcohol related HNSCC over the last few decades, there has been an increase in the number of HPV-related HNSCC (36,37). HPV+ and HPV− HNSCC differ in terms of their mutational profiles (38). Typically, HPV+ HNSCC has a better survival rate than HPV− HNSCC (36,37). Patients with HPV+ tumors have been shown to respond to chemotherapy and chemoradiation treatments at higher rates than patients with HPV− tumors, and those with HPV+ tumors have shown lower risks of progression and death than those with HPV− tumors (39).
The negative p16Ink4a status was not surprising because it is a tumor suppressor protein (40). In a normal cell, p16Ink4a inhibits cyclin dependent kinase (CDK) activity. Without p16Ink4a inhibition, CDK4 and CDK6 are able to phosphorylate the retinoblastoma tumor suppressor (Rb), and this leads to the cell cycle shifting into S phase. Without p16Ink4a there is inappropriate cell division and cell proliferation, which is why it is not surprising to find our immortalized cancer cell lines lacking p16Ink4a. However, p16Ink4a overexpression is sometimes used as a surrogate marker for high-risk-HPV-associated HNSCC (24,41,42). This is because the E7 HPV oncogene protein will induce p16Ink4a expression. The seven cell lines were identified as HPV negative, so there was no overexpression of p16Ink4a seen, as expected.
One limitation of this study is that the passage number of the cell lines is unknown. Cell lines can acquire more mutations overtime which can change their protein expression. However, a number of other labs use these cell lines too, and knowing the expression levels of MMPs and immunosuppressive biomarkers in minimal media conditions can aid in the selection of cell lines to use in future studies.
MMP and biomarker expression profiles should be carefully considered when choosing HNSCC cell lines for future studies as they can vary greatly. Because these cell lines are from different hosts, we have shown how different patients’ HNSCC cells can express varying amounts of certain biomarkers and MMPs. These differences could be due to the metastatic stage of the cancer, the primary tumor site, the type of tissue the tumor originated from, or genomic differences between patients. These results support the reason for personalized medicine and the need for further investigation into how it can be used to treat HNSCC.

Full table
Full table
Acknowledgments
Funding: This work was supported by a grant from the National Institute of Dental and Craniofacial Research (NIDCR) of the National Institutes of Health (NIH) (grant No. R01 DE014390) and a training grant from the NIDCR of the NIH (grant No. T90 DE023520). The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.05.09). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer 2011;11:9-22. [Crossref] [PubMed]
- Nagase H, Visse R, Murphy G. Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc Res 2006;69:562-73. [Crossref] [PubMed]
- Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell 2010;141:52-67. [Crossref] [PubMed]
- You TK, Kim KM, Noh SJ, et al. Expressions of E-cadherin, Cortactin and MMP-9 in Pseudoepitheliomatous Hyperplasia and Squamous Cell Carcinoma of the Head and Neck: Their Relationships with Clinicopathologic Factors and Prognostic Implication. Korean J Pathol 2012;46:331-40. [Crossref] [PubMed]
- Rabinovich GA, Gabrilovich D, Sotomayor EM. Immunosuppressive strategies that are mediated by tumor cells. Annu Rev Immunol 2007;25:267-96. [Crossref] [PubMed]
- Goncalves AS, Arantes DA, Bernardes VF, et al. Immunosuppressive mediators of oral squamous cell carcinoma in tumour samples and saliva. Hum Immunol 2015;76:52-8. [Crossref] [PubMed]
- Arantes DA, Costa NL, Mendonca EF, et al. Overexpression of immunosuppressive cytokines is associated with poorer clinical stage of oral squamous cell carcinoma. Arch Oral Biol 2016;61:28-35. [Crossref] [PubMed]
- Loffek S, Schilling O, Franzke CW. Series "matrix metalloproteinases in lung health and disease": Biological role of matrix metalloproteinases: a critical balance. Eur Respir J 2011;38:191-208. [Crossref] [PubMed]
- Khokha R, Murthy A, Weiss A. Metalloproteinases and their natural inhibitors in inflammation and immunity. Nat Rev Immunol 2013;13:649-65. [Crossref] [PubMed]
- Vilen ST, Salo T, Sorsa T, et al. Fluctuating roles of matrix metalloproteinase-9 in oral squamous cell carcinoma. ScientificWorldJournal 2013;2013:920595 [PubMed]
- Shapiro SD. Matrix metalloproteinase degradation of extracellular matrix: biological consequences. Curr Opin Cell Biol 1998;10:602-8. [Crossref] [PubMed]
- Impola U, Uitto VJ, Hietanen J, et al. Differential expression of matrilysin-1 (MMP-7), 92 kD gelatinase (MMP-9), and metalloelastase (MMP-12) in oral verrucous and squamous cell cancer. J Pathol 2004;202:14-22. [Crossref] [PubMed]
- Sutinen M, Kainulainen T, Hurskainen T, et al. Expression of matrix metalloproteinases (MMP-1 and -2) and their inhibitors (TIMP-1, -2 and -3) in oral lichen planus, dysplasia, squamous cell carcinoma and lymph node metastasis. Br J Cancer 1998;77:2239-45. [Crossref] [PubMed]
- Cathcart J, Pulkoski-Gross A, Cao J. Targeting Matrix Metalloproteinases in Cancer: Bringing New Life to Old Ideas. Genes Dis 2015;2:26-34. [Crossref] [PubMed]
- Lin CJ, Grandis JR, Carey TE, et al. Head and neck squamous cell carcinoma cell lines: established models and rationale for selection. Head Neck 2007;29:163-88. [Crossref] [PubMed]
- Brenner JC, Graham MP, Kumar B, et al. Genotyping of 73 UM-SCC head and neck squamous cell carcinoma cell lines. Head Neck 2010;32:417-26. [PubMed]
- Masters J, Palsson B. editors. Cancer cell lines. Part 2. Human cell culture v 2. London, Dordrecht: Kluwer Academic, 2002.
- Lanzel EA, Paula Gomez Hernandez M, Bates AM, et al. Predicting PD-L1 expression on human cancer cells using next-generation sequencing information in computational simulation models. Cancer Immunol Immunother 2016;65:1511-22. [Crossref] [PubMed]
- Harvey LE, Kohlgraf KG, Mehalick LA, et al. Defensin DEFB103 bidirectionally regulates chemokine and cytokine responses to a pro-inflammatory stimulus. Sci Rep 2013;3:1232. [Crossref] [PubMed]
- Borgwardt DS, Martin AD, Van Hemert JR, et al. Histatin 5 binds to Porphyromonas gingivalis hemagglutinin B (HagB) and alters HagB-induced chemokine responses. Sci Rep 2014;4:3904. [Crossref] [PubMed]
- Thunell DH, Tymkiw KD, Johnson GK, et al. A multiplex immunoassay demonstrates reductions in gingival crevicular fluid cytokines following initial periodontal therapy. J Periodontal Res 2010;45:148-52. [Crossref] [PubMed]
- Zaravinos A. An updated overview of HPV-associated head and neck carcinomas. Oncotarget 2014;5:3956-69. [Crossref] [PubMed]
- Zhao M, Sano D, Pickering CR, et al. Assembly and Initial Characterization of a Panel of 85 Genomically Validated Cell Lines from Diverse Head and Neck Tumor Sites. Clin Cancer Res 2011;17:7248-64. [Crossref] [PubMed]
- Deng Z, Hasegawa M, Aoki K, et al. A comprehensive evaluation of human papillomavirus positive status and p16INK4a overexpression as a prognostic biomarker in head and neck squamous cell carcinoma. Int J Oncol 2014;45:67-76. [Crossref] [PubMed]
- Van den Steen PE, Proost P, Wuyts A, et al. Neutrophil gelatinase B potentiates interleukin-8 tenfold by aminoterminal processing, whereas it degrades CTAP-III, PF-4, and GRO-alpha and leaves RANTES and MCP-2 intact. Blood 2000;96:2673-81. [PubMed]
- Viros D, Camacho M, Zarraonandia I, et al. Prognostic role of MMP-9 expression in head and neck carcinoma patients treated with radiotherapy or chemoradiotherapy. Oral Oncol 2013;49:322-5. [Crossref] [PubMed]
- Stanciu AE, Zamfir-Chiru-Anton A, Stanciu MM, et al. Serum Level of Matrix Metalloproteinase-9 in Patients with Head and Neck Squamous Cell Carcinoma. Clin Lab 2016;62:1569-74. [Crossref] [PubMed]
- Eckert AW, Wickenhauser C, Salins PC, et al. Clinical relevance of the tumor microenvironment and immune escape of oral squamous cell carcinoma. J Transl Med 2016;14:85. [Crossref] [PubMed]
- Shkeir O, Athanassiou-Papaefthymiou M, Lapadatescu M, et al. In vitro cytokine release profile: predictive value for metastatic potential in head and neck squamous cell carcinomas. Head Neck 2013;35:1542-50. [Crossref] [PubMed]
- Hegde S, Pahne J, Smola-Hess S. Novel immunosuppressive properties of interleukin-6 in dendritic cells: inhibition of NF-kappaB binding activity and CCR7 expression. FASEB J 2004;18:1439-41. [Crossref] [PubMed]
- Jinno T, Kawano S, Maruse Y, et al. Increased expression of interleukin-6 predicts poor response to chemoradiotherapy and unfavorable prognosis in oral squamous cell carcinoma. Oncol Rep 2015;33:2161-8. [Crossref] [PubMed]
- Aziz S, Ahmed SS, Ali A, et al. Salivary Immunosuppressive Cytokines IL-10 and IL-13 Are Significantly Elevated in Oral Squamous Cell Carcinoma Patients. Cancer Invest 2015;33:318-28. [Crossref] [PubMed]
- Ritprajak P, Azuma M. Intrinsic and extrinsic control of expression of the immunoregulatory molecule PD-L1 in epithelial cells and squamous cell carcinoma. Oral Oncol 2015;51:221-8. [Crossref] [PubMed]
- Moutsopoulos NM, Wen J, Wahl SM. TGF-beta and tumors--an ill-fated alliance. Curr Opin Immunol 2008;20:234-40. [Crossref] [PubMed]
- Benson E, Li R, Eisele D, et al. The clinical impact of HPV tumor status upon head and neck squamous cell carcinomas. Oral Oncol 2014;50:565-74. [Crossref] [PubMed]
- Rettig EM, D'Souza G. Epidemiology of head and neck cancer. Surg Oncol Clin N Am 2015;24:379-96. [Crossref] [PubMed]
- Vokes EE, Agrawal N, Seiwert TY. HPV-Associated Head and Neck Cancer. J Natl Cancer Inst 2015;107:djv344 [Crossref] [PubMed]
- Seiwert TY, Zuo Z, Keck MK, et al. Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas. Clin Cancer Res 2015;21:632-41. [Crossref] [PubMed]
- Fakhry C, Westra WH, Li S, et al. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst 2008;100:261-9. [Crossref] [PubMed]
- LaPak KM, Burd CE. The molecular balancing act of p16(INK4a) in cancer and aging. Mol Cancer Res 2014;12:167-83. [Crossref] [PubMed]
- Prigge ES, Toth C, Dyckhoff G, et al. p16(INK4a) /Ki-67 co-expression specifically identifies transformed cells in the head and neck region. Int J Cancer 2015;136:1589-99. [Crossref] [PubMed]
- Romagosa C, Simonetti S, Lopez-Vicente L, et al. p16(Ink4a) overexpression in cancer: a tumor suppressor gene associated with senescence and high-grade tumors. Oncogene 2011;30:2087-97. [Crossref] [PubMed]


