
The role of deep learning and radiomic feature extraction in cancer-specific predictive modelling: a review
Introduction
Although outcomes have improved recently, cancer is the leading cause of death in modern society (1). Personalised medicine aims to optimise outcomes by individualising cancer treatments. Numerous studies have demonstrated that extracting descriptive features of the cancer from radiological imaging data, a technique known as radiomics, can indicate the likely outcome for the patient (1-5). Radiomics is expected to be central to precision medicine due to its ability to gather detailed information describing tumour phenotypes (6).
Patient outcome measures vary depending on the type of cancer, different outcomes are important for different patients so it is important to design personalized cancer therapy treatments on a patient-by-patient basis. For example, non-small cell lung cancer (NSCLC) has been found to have poor 3-year survival, as it has a high chance of metastasizing to other organs (7). Furthermore, survival is not the only relevant outcome. The site of disease recurrence, toxicity from treatment, physical function and quality of life are all relevant in improving the cancer patient journey. Ultimately, survival is important but other outcomes worth noting are distant metastasis and local recurrence of the cancer. For some cancers, these would be considered more relevant to the success or failure of the treatment whereas a patient’s survival could be influenced by many other factors over a long follow up period.
This review explores the application of machine learning, particularly deep learning, to radiomics. Machine learning techniques can explore the relationships between images processed using radiomics, clinical outcomes and radiation dose data to help improve cancer treatment with radiotherapy. Machine learning techniques have recently been applied to medical imaging due to two main factors. The first is attributed to the significant increase in available labelled medical imaging data, the second is due to computers having increased data processing power because of parallel processing (8).
The term “radiomics” was coined by Lambin et al. (2) to describe automatic identification of unique prognostic and diagnostic features in cancer imaging data. The four main processes of radiomics include; imaging, segmentation, feature extraction and analysis, see Figure 1. It is important to note, that while Lambin et al. (2) formally raised this area, the concepts described by radiomics have been considered in many forms, many years previously through the study of tumour volumes and textures in imaging (9-11). Features are specific image characteristic (patterns) that may not be visible to a human but are recognised by a computer algorithm. Radiomics are the features that are extracted from medical images, these features can then be compared to end point data such as 2-year survival for improved prediction. The ultimate goal of radiomics is to provide a decision support system (DSS), that aids clinical decision making by providing accurate diagnosis that enables personalised radiotherapy, hopefully leading to improved treatment outcomes (3). Radiomics is primarily employed in the field of radiotherapy, however, the potential benefits of radiomics are not limited to this field.
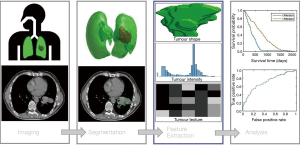
Computer vision is a general term used to describe the broad range of techniques for processing images by computers to understand or derive some useful information from digital images, these image processing techniques include pattern recognition, analysis, classification and segmentation. Computer vision may be used to automatically segment images, discover textural patterns that correlate with known materials, as well as detect and track objects in real-time. In the context of radiomics, these may correspond to tumour or organ segmentation, histopathology of cancers or real-time tumour tracking and radiotherapy delivery.
This review will focus mainly on the last two aspects of the radiomics work flow, namely feature extraction and analysis. Section I introduces the field of radiomics. Section II provides an overview of how imaging and segmentation are applied to radiomics and its importance in cancer research. Section III details the importance of feature extraction including what this is, how it has been applied in radiomics and how it has yet to be applied. Section IV details how machine learning based classification can be used to improve the analysis and prediction of cancer outcomes including how these techniques have already been applied to radiomics and what has yet to be applied to radiomics. Section V discusses how deep learning techniques may be applied to the field of radiomics by improving radiomics feature extraction and analysis of tumours. Section VI concludes this review.
Radiomic imaging and segmentation
Imaging
Imaging refers to the acquisition of a clinical image to assist in medical management. Imaging modalities include computed tomography (CT), positron emission tomography (PET), magnetic resonance imaging (MRI), and cone beam CT (CBCT). The imaging may reveal metabolic data (PET, MRI), or structural data (CT, MRI), and have variable image resolution.
CT imaging is a common, frequently used method for diagnosing, staging and following cancer. Large volumes of this imaging data are available. However, technical compatibility issues such as resolution and manufacturer’s settings impair the collation of this data into comparable forms, hence imaging data frequently requires preprocessing before analysis.
CBCT scans are acquired during treatment and suffer poorer tissue differentiation and resolution than diagnostic CT scans, however CBCT scans may be repeated frequently during treatment. Timmeren et al. (12) found that by extracting radiomic features from CBCT scans at different points during treatment, the presence or change in the delta radiomics features could be used to inform treatment options effectively.
MRI is an advanced, less frequent, more expensive technique than CT imaging which provides better tissue discrimination, multiple views, namely T1 and T2 and spectrography information which make it more likely to provide insights into cancer prognosis. Magnet strength has an impact on the quality of MRI imaging.
PET scans are obtained less frequently and have relatively poor resolution compared to CT imaging. PET scans can provide functional information about tumor metabolism, hypoxia and proliferation which are not evident from a CT.
The wide availability and use of CT imaging in diagnosis, radiotherapy planning and follow up of cancer patients makes it the preferred modality for applying radiomics and machine learning within radiotherapy as this means there is a large number of datasets available for analysis (8). Table 1 summarises the reasons for focusing on CT images. The feasibility for radiomic analysis is determined based on the image resolution and number of scans available.

Full table
Vallières et al. (13) fused FDG-PET and MRI scans in this radiomics approach with improved results, it may be that through combining multiple imaging modalities better cancer outcome prediction can be achieved.
Segmentation
Segmentation or delineation, involves selecting a part of the image for analysis, typically it contains the tumour so as to isolate the tumour signatures from the rest of the image for further analysis. Currently, the tumour volume is delineated visually by the radiation oncologist using specialised software such as Pinnacle (14). This software can provide assistance for semi-automated tumour delineation using various drawing tools and Hounsfield number-based pixel selection. Automated contouring of major organs at risk (OAR) (breast, liver, kidneys, lungs, spinal cord, brain, etc.) can now be achieved using an atlas-based segmentation approach (15,16). However, an accurate method of automatically delineating the tumour volume with no interaction from a radiation oncologist has yet to be determined for most cancer types.
Semi-automatic imaging segmentation has been compared to the macroscopic tumour seen after a surgical resection of NSCLC. Rios Velazquez et al. (4,5) uses the freely available 3D-slicer tool to delineate CT images, while Schaefer et al. (17) uses a contrast-oriented algorithm (COA) to determine the region of interest (ROI) in CT and PET images. Both of these papers report promising results in comparing the delineations to the gross tumour in the surgical specimen. Both studies utilised small sample sizes of 20 samples for Velazquez et al. (5) and 15 samples for Schaefer et al. (17). Haga et al. (18) also studied the problem of inter-observer variability in their study of 40 NSCLC patients where a semi-automatic gross tumour volume (GTV) contour is compared to the observations from three oncologists.
Sun et al. (19) have recently used Gaussian kernel support vector machines (SVM) to predict the tumour location in prostate cancer with high results. Deep learning has also been applied to this problem of atlas-based segmentation of breast and brain cancer (20,21) with success, to extract learned imaging features and then train a classifier to predict the tumour location. This area is a growing field, which has vast scope for improvement. For the purposes of this review, only papers that analyse the GTV defined by an oncologist will be considered as the ground truth in analyzing the tumour volume via radiomics.
Radiomic feature extraction
Feature extraction in radiomics
It is important to note the difference between feature extraction and feature selection. The goal of feature extraction is to find as many features as possible that describe the collected data. The purpose of feature selection is to reduce the multitude of extracted features to as few as possible, so that these in turn can be generalised as patterns that robustly identify the concepts hidden within the data, while at the same time avoiding over-fitting the data. Over-fitting the data is a problem in machine learning where the analysis may have excellent results when applied to the training data, but when new data is presented for analysis, the model has a degraded performance.
Feature selection is currently the main method for determining radiomic signatures, and is commonly used in the literature (1,2,13,22,23). Additionally, Ypsilantis et al. (24) has employed Convolutional Neural Networks (CNN) for feature extraction in the field of radiomics.
There are numerous dimensionality reduction methods for selecting and minimising the number of features once found. These include Principal Component Analysis (PCA) (10,25,26), covariance matrices or linear discriminant analysis (LDA) (27-29).
The previous proven method of deterministic feature extraction was based on Haralick textural feature analysis (9). There are two main approaches for feature extraction: deterministic and nondeterministic extraction. Deterministic feature extraction is the most common method where a mathematical formula is employed to extract features relating to imaging features such as texture, intensity or shape, this method is employed in the following papers (1,22,23,28,30-34).
The current literature suggests that texture features are the one of the most important imaging features for the field of radiomics (1,13,35), hence state-of-the-art texture detection computer vision algorithms will be explored in detail in section IV. Sollini et al. (35) extensive review of the literature yielded that PET CT imaging is crucial for the analysis of cancer data due to the additional information it can provide relating to tumour staging and response, with textural radiomics features being the most prognostic.
The application of feature extraction techniques to radiomics
The feature extraction process utilises a variety of image processing techniques and mathematical analyses to find common patterns within the images. It has been found that texture features are the one of the most important imaging features for the field of radiomics (1-5,13,17), hence state-of-the-art texture detection computer vision algorithms will be explored in detail in section IV and V.
Kumar et al. (36) describe that there are a vast number of extractible radiomic features for each patient analysed, hence the quantity of features needs to be reduced to avoid overfitting the data. They suggest that only 39 of their 327 quantitative features found were reproducible, informative and not redundant. The extraction of features in this paper required applying a statistical formula or imaging filter to determine the radiomic features. They found the ability to relate a specific feature to a specific cancer type or outcome is very difficult to achieve.
The relationship between radiomic features and the prediction of cancer survival rates, was examined in Aerts et al. (1) who extracted 440 radiomic features from four main categories (I: texture, II: wavelet or image transformations, III: shape and IV: tumour intensity), but ultimately found that only four radiomic features (absence or presence) were needed to determine a patient’s likelihood of 2-year survival with moderately high accuracy (1). Many other authors have undertaken the extraction of various radiomics features from images to determine if there is a correlation between these mathematically determined features and survival, and many have found recurring features, also strongly correlated with survival (3,13,22,23,28,30,36-38).
Vallières et al. (13) introduced a novel radiomics approach by fusing FDG-PET scans with MRI imaging to accurately predict the occurrence of lung metastasis in patients with soft tissue sarcoma (STS). Even with data loss with low resolution FDG-PET scans relative to MRI images, the fusion of these two modalities allowed the texture and shape of the tumour to be clearly highlighted and clearly predicted patients who developed lung metastasis compared to those who would not. This allowed for the discovery of new radiomics features, which were found to be strongly correlated with patient survival outcomes. They found that the maximum Standardized Uptake Value (SUVmax) was significantly associated with lung metastasis risk, which is not unexpected since this is used to measure the tumour size and aggression. Other radiomics studies (39-41) had independently determined that SUVmax is strongly correlated with tumour heterogeneity. The development of lung metastasis portends a very low 3-year survival rate, so the group predicted to have lung metastases can be defined as being at high risk of recurrence. It is then possible to apply additional treatments to this group to see if the incidence of lung metastasis recurrence can be lowered.
Mi et al. (42) introduced a robust feature selection technique known as hierarchical forward selection (HFS) in the hope of overcoming the issue of small sample sizes faced by medical imaging datasets. Where an SVM is employed to evaluate each of the features obtained and only retains features that are strongly correlated with improved prediction accuracy. This method proved to be highly accurate when applied to PET images.
Feature extraction techniques yet to be applied to radiomics
The Fisher vector (FV) and 3D shape invariant feature transform (3D SIFT) have been combined with CNN to perform traditional machine learning based classification for improved feature extraction (43,44). Sánchez et al. (45) introduce FV as an alternative supervised learning classification technique based on Gaussian mixture modelling (GMM) that has been found to offer advanced competitiveness in classifying images. The FV is based on the Fisher kernel patch encoding technique, which uses GMM to describe an image as a finite number of clusters. This is an alternative to the comparable popular representation, the bag of visual words (BoV). The FV is a high dimensional vector which has been found to have high accuracy and speed for classifying various image datasets with over 8 million images (45). This method employs product quantization to improve efficiency and accuracy by reducing the data with negligible losses.
The FV is computed by, separating the image into windowed voxels of the tumour. The size of the window may be varied experimentally to determine the window size that achieves optimal results. Local features can be calculated using radiomic analysis, such as a gray-level co-occurrence matrix (GLCM) or logarithmic features, which have been found to be optimal for predicting distant metastasis in lung adenocarcinomas by Coroller et al. (23).
3D SIFT presented by Scovanner et al. (46) may be employed as a feature descriptor for 3D images to interpret the words (areas) in the 3D tumour image into biometric signatures. These signatures can be utilised for improved efficient linear classification. Paganelli et al. (47) have successfully employed SIFT features for tracking tumours in 4DCT scans and for image registration of CBCT scans.
Radiomic analysis using machine learning
Machine learning
Machine learning can be used to determine which of a plethora of features alone or in combination are strongly correlated with outcomes for any cancer type. More importantly, machine learning techniques such as deep learning and other neural networks allow for the discovery of relationships that have not been considered within the radiomic feature set extracted. Machine learning can be separated into three categories: supervised, unsupervised and semi-supervised learning.
Supervised learning requires a labelled imaging and outcome dataset that may be used to find the image representation features that most closely relate to the classification or outcome. Conversely, unsupervised learning does not involve a labelled imaging dataset but does require an outcome; in this case one is looking for image representation features that best describe the image as a whole. Semi-supervised learning involves a labelled imaging and outcome dataset which may be missing information that can be inferred to enable an accurate classification. Supervised and unsupervised learning techniques are often combined to achieve superior results.
Supervised feature extraction starts with a defined feature and determines if it is significant for predicting ground truth data such as survival outcome. Unsupervised feature extraction involves a machine learning method, whether deep learning or clustering, to extract textual features that form repeatable models of sub concepts in the data, before determining if any of these discovered features predict ground truth data such as survival outcome. Unsupervised feature extraction is ideal when the useful, important features of a dataset are unknown.
The application of machine learning to radiomics
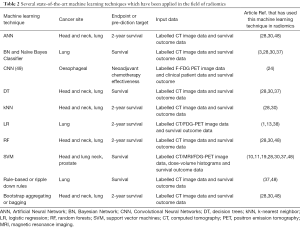
Table 2 summarises the advantages and disadvantages of various machine learning techniques that have already been applied to the field of radiomics. The machine learning techniques listed in this table are compared and discussed in this section. Machine learning has been rapidly applied to radiomics, but it is important to note that these techniques focus mainly on analysing the radiomic features compiled by other authors in the field (1,13,36), but they do not focus on using deep learning to add to these known features.
Full table
The analysis process includes utilizing the discovered features in a classification model to predict the likely outcome using classifiers such as decision trees (DT), random forests (RF) or GMM to predict important patient information including 2-year survival or likelihood of cancer spread. DT are a machine learning technique that separates outcomes based on the statistical significance, displayed as a probability tree. DT are ideal for classification as they can identify and provide additional contextual information to an oncologist to support and improve their decision making by using medical expertise to justify the prediction. Modelling can help inform experts about disease processes which they can match with their clinical expertise.
Radiomic features extracted from imaging data, tumour volume and radiation dose data (all available in a digital imaging and communications in medicine-radiation therapy (DICOM-RT) standard file describing radiotherapy treatment) can be combined with other visual data such as histopathological images (1), to correlate with outcome data, whether survival or toxicity. Observed patient outcomes are used in the training data set to establish a machine learning model which is able to predict outcomes of patient treatments. The tumour volume is used as it represents the domain expert’s visual conclusion about tumour location.
A promising radiomics-based lung cancer DSS was developed by Dekker et al. (3) The DSS was based on routine care data in an attempt to personalise decisions about appropriate curative or palliative radiotherapy doses based on outcomes in NSCLC. This study analysed 322 patients from the Netherlands in the training cohort and 159 patients from Australia in the clinical cohort, using clinical data (tumour volume, radiotherapy dose, lung function, age, gender) to predict 2-year survival. The DSS separated the patients with high accuracy (P<0.001) into 2 groups—a poor/medium prognosis group and a good prognosis group. In the course of this study, it was found that Bayesian Networks (BN) produced superior results for the imputation of missing features, as they can be used to impute missing data by inferring from existing data. This is useful in clinical data analysis, since it is common for medical databases to contain missing data.
The Dekker DSS (3) provided the useful clinical finding, that patients classified as “good prognosis” do poorly if treated with palliative radiotherapy (20–30 Gy in 1–2 weeks) but well if treated with curative radiotherapy (60 Gy in 6 weeks). The difference was a 40% improvement in 2-year survival. Secondly, that “poor/medium prognosis” patients do poorly whether treated with curative radiotherapy or palliative radiotherapy. The ability to classify lung cancer patients allows the survival of “good prognosis” patients to be optimised, and the time of “poor/medium prognosis” not to be wasted on lengthy unhelpful radiotherapy.
Machine learning techniques such as BN, Neural Networks (NN), DT and SVM have been employed in numerous studies to predict 2-year survival (3,28,30,37). The majority of these studies have chosen to use SVM, which are limited to providing only two classification options. This output is adequate to predict a patient’s likelihood of 2-year survival, however other machine learning techniques such as DT can produce more detailed informative analysis of medical images.
A promising early radiomics study by El Naqa et al. (10) used PCA to uncover non-linear behaviour, which was then combined with a SVM kernel method to achieve superior results in predicting disease endpoints. This approach validates the potential for statistical learning in radiotherapy to improve personalised treatment outcomes.
Parmar et al. (28) investigated various machine learning and feature selection techniques for NSCLC cancer patients. This paper compared twelve machine learning techniques, including but not limited to, NN, DT, SVM, RF, kNN and LDA. In this study, RF were found to have the highest overall accuracy for determining 2-year survival. The study included a 310-patient training cohort and a 154-patient validation cohort. Unsupervised learning techniques, such as clustering or self organising maps (SOM), have better performance when very large datasets are provided. Larger cohorts of patients will improve the credibility of this applied research to reliably predict 2-year survival in NSCLC patients.
Hawkins et al. (37) compared four machine learning techniques including DT, Rule Based Classification, Naive Bayes and SVM with various feature selection techniques such as test-retest, relief-F and correlation-based feature selection. This study included 81 lung patients with adenocarcinoma (one histopathological type of NSCLC) in the training cohort, and concluded that the decision tree once validated with a leave-one-out-cross validation approach obtained the best accuracy for predicting 2-year survival. Two key draw backs of this study are the small dataset and the lack of separation between training and validation datasets.
Machine learning techniques yet to be applied to radiomics
There are many alternative classification methods in machine learning that are yet to be explored fully through radiomics analysis such as DT, SVM and LR. Their ability to perform well in radiomics depends more on the type of output required. SVM and LR are useful in separating cohorts into two different groups, for example good and bad prognosis, but they fail to provide further information. DT can be used to separate cohorts into multiple groups, and the results are completely interpretable, allowing an expert to understand and implement the result in the treatment of patients.
For future research in radiomics, suitability for distributed learning is an essential criterion. Radiomics requires large amounts of data to develop robust models; this volume of data will need to be collected from multiple hospitals in multiple state and national jurisdictions in order to achieve a generic model which can be applied in all situations to support personalised cancer therapy. Lambin et al. (50) stresses the need for a distributed DSS where instead of sharing data, institutions may share a model to describe the data such as the result from a supervised machine learning technique. The models from each individual institution can then be combined to form a consensus with higher accuracy (51,52). On the other hand, Skripcak et al. (53) discussed the implementation of a federated international database containing thousands of de-identified patient data sets from multiple institutions around the world to serve the same purpose. This type of international database would be ideal for unsupervised learning techniques such as CNN.
Some unsupervised classification methods yet to be applied to radiomics include variational Bayesian Gaussian mixture modelling (VB-GMM) and minimum message length (MML). VB-GMM has been proven to perform better linear classification than BoV, thereby making it far more suited to distributed learning (54).
Clustering is another form of unsupervised learning used to categorise and analyse data. Clustering techniques, such as k-means, GMM or FVs, can be used to divide the tumour into windows or regions grouped by similarity of the image features. These clustered areas can then be classified into different clinical tissue types, such as necrotic tissue, normal tissue and proliferating/aggressive tumours. Using clustering to locate regions of interest within a tumour allows for the identification of normal tissue to assist with reducing normal tissue toxicity by identifying regions to be avoided during radiotherapy such as OAR. Even et al. (55) successfully employed clustering using mutli-parametric functional imaging to detect metabolic activity (FDG-PET/CT) and hypoxia (HX4-PET/CT), which allowed them to group regions into phenotypic clusters which found that clusters that were highly hypoxic and metabolic had poorer survival outcomes.
GMM may be used to cluster features into a finite vocabulary. This is a key difference between the BoV approach and the FV as the BoV method employs k-means clustering instead of GMM. The main difference between k-means and GMM is in how membership to clusters is assigned. K-means assigns data points to a cluster based on the minimum (Euclidean) distance. GMM uses a “soft” probabilistic assignment of a data point to the cluster. The feature vector generated from GMM calculates two very important GMM variables, the mean and variance, for each windowed region, the regions are then clustered based on closest means and furthest variance. This ensures that the clusters similarities and differences are accentuated so that the clusters are as precise as possible.
GMM or k-means may be used to compute the clusters into a finite vocabulary a histogram can be generated to form words (areas) inside the tumour region. Ideally, this form of recurring patterns or motifs can be extracted for classification purposes.
Supervised classification may be employed to predict the overall survival likelihood model from the resulting FV, through comparison with outcome data.
Clinical perspective for clustering
Prise et al. (56) reviewed the state-of-the-art research in radiotherapy by analysing the “by-stander” effect of radiotherapy on healthy tissue. The by-stander effect occurs when cells, which were not irradiated, receive a signal from an irradiated cell that leads to the un-irradiated cell dying, thereby producing unforeseen tissue damage. The authors suggest that future studies should focus on using multiple small beams to irradiate a tumour rather than using a uniform large beam of the target area, in order to minimise damage to healthy tissue.
The prevailing radiomics techniques can be applied to regions clustered around the irradiated regions, attempting to find textural features and radiomic signatures that may be related to likelihood of cell death. The comparison of clustered regions with genomic data, might indicate that certain clusters have a different prognosis depending on the patient’s genes. This would allow greater personalisation of treatment with the hope of improved patient outcomes.
Radiomic analysis using deep learning
This section will explore state-of-the-art deep learning techniques such as CNN, Deep Belief Networks (DBN) and Deep Autoencoders which have the ability to service radiomics as they can detect textures in images. It is important to note that deep learning is another form of machine learning, the potential for deep learning in radiomics will be explained in this section.
Deep learning
Research applications in medical imaging using deep learning have increased recently due to the amount of available medical imaging data increasing, and also because parallel processing by computers have allowed increased computation speeds. However, a main hurdle in applying deep learning to medical imaging remains, namely that deep learning requires extensive amounts of labelled medical data and this is difficult and time consuming to obtain within the medical field (8). Another issue is that experts can sometimes contradict each other. Despite this CNNs show the most promise in extracting features from medical images for classification especially for the image registration or segmentation of tumours (57).
There are many different deep learning architectures or methods including but not limited to; DNN, Deep Autoencoders, DBN, Deep Boltzmann Machines (DBM), Recurrent Neural Networks (RNN) and CNN. The majority of these architectures perform better with a single data stream of data rather than imaging data, and so are unsuitable for medical imaging applications. However, CNN are ideal for analysis of 2D imaging data. Recently CNN has been successfully used to analyse 3D imaging data, which is important, as tumour detection and delineation is a 3D imaging problem. It is therefore essential to utilise machine learning techniques that can handle this type of data well (8).
A recent paper by LeCun et al. (49) provides a detailed review of how deep learning is revolutionizing computer vision through the use of CNN. CNN is a biologically influenced technique inspired by how the human brain translates its own visual inputs; it was originally developed in the late 90s but was considered too slow for efficient pattern recognition. However, with the advancement of fast computing, this technique has now been found to be superior to other traditional supervised machine learning techniques in interpreting and classifying raw image data. The conventional supervised machine learning techniques struggle to process raw data in one step as the data needs to be firstly pre-processed, then features extracted and selected using mathematical models, and then finally prediction algorithms can be applied. In addition to this expert knowledge of the data is often required to interpret such features correctly, which is very time consuming.
CNNs are feed-forward networks that have been applied to image processing with impressive results and can be employed to process information for 3D video or volumetric images, which equips them as promising feature selection technique for volumetric tumour imaging data. This technique is limited in that it requires a large amount of processing power and is highly dependent on the information that is fed into the training algorithm. For this reason, the training data must be a good representation of all the different types of outcomes, to avoid over-fitting.
The process for creating a CNN has three main steps. Firstly, the convolutional layer is formed where the input data or image is convolved into several small kernels or filters. Secondly, the pooling layer is formed where the resulting kernels from the previous step are down-sampled, often via a max pooling process. This involves separating the image into regions with the highest kernel value from each region being selected and conveyed to the next step. These first two steps combined will produce a one-layer CNN. The third step involves repeating the first and second steps, with the output from the second step serving as input to the first step.
The key benefit of CNNs is that despite the lengthy training time, they can classify images extremely quickly as the features that are extracted are simple non-linear functions and convolutions (58). The first layer often detects primitive patterns such as edges; the next layer finds larger patterns and so on. Higher layer CNNs can often produce higher level features similar to the features extracted in the field of radiomics. In this way a CNN can be used to convert 3D images into 1D vectors to allow formal classification through conventional machine learning methods (8).
There is a question about how much data is required to produce useful results in deep learning. While most deep learning applications such as CNN require very large data sets in the order of thousands of examples, this does not necessarily imply thousands of patients are required to produce useful results. In the case of radiomics, this involves very large radiology imaging data sets which may only have a few hundred individual patients. In these cases, CNN can be employed to segment the radiology images into smaller patches (sub-images or voxels) that can be used to train and classify the data, thereby supplying hundreds of thousands of images from only a few hundred patients. Recent deep learning studies have involved the use of CNN on radiology, histopathologic and cell images for less than 100 patients, including the validation data set, and produced superior classification results (43,44,59).
The application of deep learning to radiomics
DBN are often combined with CNN, to create Convolutional Deep Belief Networks (CDBN) that model the data more effectively. Hinton et al. (60) developed an advanced learning algorithm for DBN which integrated with CNN. The DBN methodology develops a generative model of multiple tiers that finds statistical similarities between tiers to increase the speed of overall training while maintaining data integrity. It is likely that CDBN may provide improved results for image analysis in radiomics (8).
Lee et al. (61) and Wu et al. (62) have combined CNN and DBN with promising results. Lee and colleagues (61) introduce a technique known as “probabilistic max-pooling” that reduces the data to be analysed and thereby increases the efficiency, and demonstrated high accuracy for multiple pattern recognition tasks for large image sets (61). Wu et al. (62) avoided max-pooling on a CDBN to recognize 3D shapes on a 3D voxel grid, as it increased the uncertainty of shape reconstruction for pattern recognition of 3D point cloud images, and were able to recognize 3D shapes with high precision and efficiency from low quality input data with state-of-the-art performance in numerous tasks. CDBN have also been applied successfully to medical imaging tasks (63,64).
Deep Autoencoders have been applied in medical imaging for feature extraction through data driven learning (65,66). Since this method employs unsupervised learning, labelled imaging data is not required for training. There are variants of Autoencoders, and the Convolutional Autoencoder is the main type that is likely to be useful for radiomics. A Convolutional Autoencoder preserves spatial locality and can be applied to 2D images (8).
Radiomics is a growing field that is based on the analysis of hand-crafted features which depend on an arbitrary decision to apply a statistical analysis to an image, as a form of feature engineering. Deep learning has the potential to extract learned features from images which may be more useful in determining the required outcome. By combining the learned features extracted via deep learning along with the current hand-crafted radiomic features it may be possible to improve outcome prediction. Machine learning techniques such as DTs may also be useful in determining which learned features are most prognostic. Deep learning can also be employed to segment the tumour out from the image. Several authors have already looked into applying deep learning to radiomics with improved results, leading to a higher potential for personalised cancer treatment (24,67). Li et al. (67) employed a six-layer CNN to extract features along with the FV to classify the features and found an improvement on the traditional radiomics method obtaining 95% accuracy compared to 86%.
Limitations of deep learning in medical imaging
The availability of open source deep learning packages (68-77) has led to its rapid adoption in the field of medical imaging, however deep learning still has limitations in the field of radiomics and medical imaging. In order to interpret and apply these deep learning effectively significant expertise in biology and computer science is required (8). This is due to the black box nature of deep learning where a result is generated with high model accuracy with no specific medical based reason. Hence, the results from deep learning can be difficult to interpret clinically, which can limit their use in decision making. However, deep learning is useful for generating radiomic features which can then be interpreted using another machine learning method such as DT. DTs can show how the results were generated, which factors are most important in influencing the final result, and lend themselves to being validated by clinical trials. DTs also permit the inclusion of other medical data (diagnosis, stage, grade, etc.) to aid in interpreting the results obtained through deep learning (8).
Deep learning tends to suffer from the issue of over-fitting, where it performs well with minimal errors on the original training data. However, the inherent model learnt can be overly specific and not representative of the underlying concepts captured in the data. Hence, when new data is introduced to the over-fitted model, the model struggles to perform well. This problem can be solved by introducing a larger and more representative training set that models the population more accurately.
Conclusions
The potential of radiomics as a clinical tool exists wherever radiotherapy is used with contemporaneous imaging. Modern planning uses CTs, PETs, CBCTs and MRIs. The MRI-Linac is the state-of-the-art device which allows a tumour to be MRI-scanned and irradiated at the same time (78). Combining radiomics with machine learning through the implementation of real-time image processing, analysis and identification of cancer will support clinical decision making, possibly during the delivery of radiotherapy through technologies such as the MRI-Linac (79).
Radiomics is a rapidly advancing field of clinical image analysis with a vast potential for supporting decision making involved in the diagnosis and treatment of cancer (80). The realisation of this goal of more effective decision making requires significant individual and integrated expertise from domain experts in medicine, biology and computer science to allow advances in computer vision and machine learning techniques to be applied effectively. Deep learning combined with Machine learning has the potential to advance the field of radiomics significantly in the years to come provided the raw data is made available for the results to be determined robustly across all patient and tumour types (81).
Acknowledgments
Funding: This research has been conducted with the support of the Australian Government Research Training Program Scholarship, along with the Radiation Oncology Staff Specialist Trust Funds at Illawarra Cancer Care Centre, Wollongong, NSW, Australia and the Liverpool Cancer Therapy Centre, Liverpool, NSW, Australia.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.05.02). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. [Crossref] [PubMed]
- Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441-6. [Crossref] [PubMed]
- Dekker A, Vinod S, Holloway L, Oberije C, et al. Rapid learning in practice: A lung cancer survival decision support system in routine patient care data. Radiother Oncol 2014;113:47-53. [Crossref] [PubMed]
- Rios Velazquez E, Aerts HJ, Gu Y, et al. A semiautomatic CT-based ensemble segmentation of lung tumors: Comparison with oncologists’ delineations and with the surgical specimen. Radiother Oncol 2012;105:167-73. [Crossref] [PubMed]
- Velazquez ER, Parmar C, Jermoumi M, et al. Volumetric CT-based segmentation of NSCLC using 3D-Slicer. Sci Rep 2013;3:3529. [Crossref] [PubMed]
- Chen B, Zhang R, Gan Y, et al. Development and clinical application of radiomics in lung cancer. Radiat Oncol 2017;12:154. [Crossref] [PubMed]
- Arrieta O, Villarreal-Garza C, Zamora J, et al. Long-term survival in patients with non-small cell lung cancer and synchronous brain metastasis treated with whole-brain radiotherapy and thoracic chemoradiation. Radiat Oncol 2011;6:166. [Crossref] [PubMed]
- Ravi D, Wong C, Deligianni F, et al. Deep Learning for Health Informatics. IEEE J Biomed Health Inform 2017;21:4-21. [Crossref] [PubMed]
- Haralick RM, Shanmugam K, Dinstein I. Textural Features for Image Classification. IEEE Trans Syst Man Cybern 1973;SMC-3:610-21. [Crossref]
- El Naqa I, Bradley JD, Lindsay PE, et al. Predicting radiotherapy outcomes using statistical learning techniques. Phys Med Biol 2009;54:S9-30. [Crossref] [PubMed]
- Dehing-Oberije C, Yu S, De Ruysscher D, et al. Development and External Validation of Prognostic Model for 2-Year Survival of Non-Small-Cell Lung Cancer Patients Treated With Chemoradiotherapy. Int J Radiat Oncol Biol Phys 2009;74:355-62. [Crossref] [PubMed]
- van Timmeren JE, Leijenaar RT, van Elmpt W, et al. Feature selection methodology for longitudinal cone-beam CT radiomics. Acta Oncol 2017;56:1537-43. [Crossref] [PubMed]
- Vallières M, Freeman CR, Skamene SR, et al. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys Med Biol 2015;60:5471. [Crossref] [PubMed]
- Mitchell RA, Wai P, Colgan R, et al. Improving the efficiency of breast radiotherapy treatment planning using a semi-automated approach. J Appl Clin Med Phys 2017;18:18-24. [PubMed]
- Daisne JF, Blumhofer A. Atlas-based automatic segmentation of head and neck organs at risk and nodal target volumes: A clinical validation. Radiat Oncol 2013;8:154. [Crossref] [PubMed]
- Bell LR, Dowling JA, Pogson EM, et al. Atlas-based segmentation technique incorporating inter-observer delineation uncertainty for whole breast. J Phys Conf Ser 2017;777:012002-1-012002-4. [Crossref]
- Schaefer A, Kim YJ, Kremp S, et al. PET-based delineation of tumour volumes in lung cancer: comparison with pathological findings. Eur J Nucl Med Mol Imaging 2013;40:1233-44. [Crossref] [PubMed]
- Haga A, Takahashi W, Aoki S, et al. Classification of early stage non-small cell lung cancers on computed tomographic images into histological types using radiomic features: interobserver delineation variability analysis. Radiol Phys Technol 2018;11:27-35. [Crossref] [PubMed]
- Sun Y, Reynolds H, Wraith D, et al. Predicting prostate tumour location from multiparametric MRI using Gaussian kernel support vector machines. Australas Phys Eng Sci Med 2017;40:39-49. [Crossref] [PubMed]
- Petersen K, Nielsen M, Diao P, et al. Breast tissue segmentation and mammographic risk scoring using deep learning. In: Fujita H, Hara T, Muramatsu C, et al. editors. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). Berlin: Springer International Publishing, 2014.
- Işin A, Direkoǧlu C, Şah M. Review of MRI-based Brain Tumor Image Segmentation Using Deep Learning Methods. Procedia Comput Sci 2016;102:317-24. [Crossref]
- Leijenaar RT, Nalbantov G, Carvalho S, et al. The effect of SUV discretization in quantitative FDG-PET Radiomics: the need for standardized methodology in tumor texture analysis. Sci Rep 2015;5:11075. [Crossref] [PubMed]
- Coroller TP, Grossmann P, Hou Y, et al. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother Oncol 2015;114:345-50. [Crossref] [PubMed]
- Ypsilantis PP, Siddique M, Sohn HM, et al. Predicting Response to Neoadjuvant Chemotherapy with PET Imaging Using Convolutional Neural Networks. PLoS One 2015;10:e0137036 [Crossref] [PubMed]
- Kickingereder P, Burth S, Wick A, et al. Radiomic Profiling of Glioblastoma: Identifying an Imaging Predictor of Patient Survival with Improved Performance over Established Clinical and Radiologic Risk Models. Radiology 2016;280:880-9. [Crossref] [PubMed]
- Larue RT, Defraene G, De Ruysscher D, et al. Quantitative radiomics studies for tissue characterization: a review of technology and methodological procedures. Br J Radiol 2017;90:20160665 [Crossref] [PubMed]
- Parmar C, Leijenaar RT, Grossmann P, et al. Radiomic feature clusters and Prognostic Signatures specific for Lung and Head & Neck cancer. Sci Rep 2015;5:11044. [Crossref] [PubMed]
- Parmar C, Grossmann P, Bussink J, et al. Machine Learning methods for Quantitative Radiomic Biomarkers. Sci Rep 2015;5:13087. [Crossref] [PubMed]
- Balagurunathan Y, Kumar V, Gu Y, et al. Test-Retest Reproducibility Analysis of Lung CT Image Features. J Digit Imaging 2014;27:805-23. [Crossref] [PubMed]
- Parmar C, Grossmann P, Rietveld D, et al. Radiomic Machine-Learning Classifiers for Prognostic Biomarkers of Head and Neck Cancer. Front Oncol 2015;5:272. [Crossref] [PubMed]
- Balagurunathan Y, Gu Y, Wang H, et al. Reproducibility and Prognosis of Quantitative Features Extracted from CT Images. Transl Oncol 2014;7:72-87. [Crossref] [PubMed]
- Cunliffe A, Armato SG, Castillo R, et al. Lung Texture in Serial Thoracic Computed Tomography Scans: Correlation of Radiomics-based Features With Radiation Therapy Dose and Radiation Pneumonitis Development. Int J Radiat Oncol Biol Phys 2015;91:1048-56. [Crossref] [PubMed]
- Leijenaar RT, Carvalho S, Hoebers FJ, et al. External validation of a prognostic CT-based radiomic signature in oropharyngeal squamous cell carcinoma. Acta Oncol 2015;54:1423-9. [Crossref] [PubMed]
- Zhu Y, Li H, Guo W, et al. Deciphering Genomic Underpinnings of Quantitative MRI-based Radiomic Phenotypes of Invasive Breast Carcinoma. Sci Rep 2015;5:17787. [Crossref] [PubMed]
- Sollini M, Cozzi L, Antunovic L, et al. PET Radiomics in NSCLC: State of the art and a proposal for harmonization of methodology. Sci Rep 2017;7:358. [Crossref] [PubMed]
- Kumar V, Gu Y, Basu S, et al. Radiomics: The process and the challenges. Magn Reson Imaging 2012;30:1234-48. [Crossref] [PubMed]
- Hawkins SH, Korecki JN, Balagurunathan Y, et al. Predicting Outcomes of Nonsmall Cell Lung Cancer Using CT Image Features. IEEE Access 2014;2:1418-26.
- Vial A, Stirling D, Field M, et al. Assessing the prognostic impact of 3D CT image tumour rind texture features on lung cancer survival modelling. Montreal: 2017 IEEE Global Conference on Signal and Information Processing (GlobalSIP), 2017:735-9.
- Ganeshan B, Skogen K, Pressney I, et al. Tumour heterogeneity in oesophageal cancer assessed by CT texture analysis: Preliminary evidence of an association with tumour metabolism, stage, and survival. Clin Radiol 2012;67:157-64. [Crossref] [PubMed]
- Rahmim A, Schmidtlein CR, Jackson A, et al. A novel metric for quantification of homogeneous and heterogeneous tumors in PET for enhanced clinical outcome prediction. Phys Med Biol 2016;61:227-42. [Crossref] [PubMed]
- Lin P, Min M, Lee M, et al. Nodal parameters of FDG PET/CT performed during radiotherapy for locally advanced mucosal primary head and neck squamous cell carcinoma can predict treatment outcomes: SUVmean and response rate are useful imaging biomarkers. Eur J Nucl Med Mol Imaging 2017;44:801-11. [Crossref] [PubMed]
- Mi H, Petitjean C, Dubray B, et al. Robust feature selection to predict tumor treatment outcome. Artif Intell Med 2015;64:195-204. [Crossref] [PubMed]
- Stanitsas P, Cherian A, Li X, et al. Evaluation of feature descriptors for cancerous tissue recognition. 2016 23rd International Conference on Pattern Recognition, ICPR 2016. United States: Institute of Electrical and Electronics Engineers Inc, 2017:1490-5.
- Gao Z, Wang L, Zhou L, et al. HEp-2 Cell Image Classification With Deep Convolutional Neural Networks. IEEE J Biomed Health Inform 2017;21:416-28. [Crossref] [PubMed]
- Sánchez J, Perronnin F, Mensink T, et al. Image Classification with the Fisher Vector: Theory and Practice. Int J Comput Vis 2013;105:222-45. [Crossref]
- Scovanner P, Ali S, Shah M. A 3-dimensional Sift Descriptor and Its Application to Action Recognition. Proceedings of the 15th ACM International Conference on Multimedia. New York, NY, USA: ACM, 2007:357-60.
- Paganelli C, Peroni M, Pennati F, et al. Scale Invariant Feature Transform as feature tracking method in 4D imaging: a feasibility study. Conf Proc IEEE Eng Med Biol Soc 2012;2012:6543-6. [PubMed]
- de Carvalho Filho AO, Silva AC, de Paiva AC, et al. Lung-Nodule Classification Based on Computed Tomography Using Taxonomic Diversity Indexes and an SVM. J Signal Process Syst 2016;87:179-96. [Crossref]
- LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436-44. [Crossref] [PubMed]
- Lambin P, Zindler J, Vanneste BG, et al. Decision support systems for personalized and participative radiation oncology. Adv Drug Deliv Rev 2017;109:131-53. [Crossref] [PubMed]
- Jochems A, Deist TM, El Naqa I, et al. Developing and Validating a Survival Prediction Model for NSCLC Patients Through Distributed Learning Across 3 Countries. Int J Radiat Oncol Biol Phys 2017;99:344-52. [Crossref] [PubMed]
- Deist TM, Jochems A, van Soest J, et al. Infrastructure and distributed learning methodology for privacy-preserving multi-centric rapid learning health care: euroCAT. Clin Transl Radiat Oncol 2017;4:24-31. [Crossref] [PubMed]
- Skripcak T, Belka C, Bosch W, et al. Creating a data exchange strategy for radiotherapy research: Towards federated databases and anonymised public datasets. Radiother Oncol 2014;113:303-9. [Crossref] [PubMed]
- Valente F, Wellekens CJ. Variational Bayesian GMM for speech recognition. Geneva, Switzerland: EUROSPEECH 2003, 8th European conference on speech communication and technology, 2003.
- Even AJ, Reymen B, La Fontaine MD, et al. Clustering of multi-parametric functional imaging to identify high-risk subvolumes in non-small cell lung cancer. Radiother Oncol 2017;125:379-84. [Crossref] [PubMed]
- Prise KM, O’Sullivan JM. Radiation-induced bystander signalling in cancer therapy. Nat Rev Cancer 2009;9:351-60. [Crossref] [PubMed]
- Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60-88. [Crossref] [PubMed]
- McCann MT, Jin KH, Unser M. Convolutional Neural Networks for Inverse Problems in Imaging: A Review. IEEE Signal Process Mag 2017;34:85-95. [Crossref]
- Huang X, Shan J, Vaidya V. Lung nodule detection in CT using 3D convolutional neural networks. Melbourne, Australia: 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), 2017:379-83.
- Hinton GE, Osindero S, Teh YW. A Fast Learning Algorithm for Deep Belief Nets. Neural Comput 2006;18:1527-54. [Crossref] [PubMed]
- Lee H, Grosse R, Ranganath R, et al. Convolutional deep belief networks for scalable unsupervised learning of hierarchical representations. In: Proceedings of the 26th Annual International Conference on Machine Learning SE - ICML ’09. New York, NY, USA: ACM, 2009:609-16.
- Wu Z, Song S, Khosla A, et al. 3D ShapeNets: A Deep Representation for Volumetric Shapes. Available online: http://arxiv.org/abs/1406.5670
- Zhen X, Wang Z, Islam A, et al. Multi-scale deep networks and regression forests for direct bi-ventricular volume estimation. Med Image Anal 2016;30:120-9. [Crossref] [PubMed]
- Brosch T, Tam R. Manifold Learning of Brain MRIs by Deep Learning BT - Medical Image Computing and Computer-Assisted Intervention - MICCAI 2013. In: Mori K, Sakuma I, Sato Y, editors. Berlin, Heidelberg: Springer, 2013:633-40.
- Avendi MR, Kheradvar A, Jafarkhani H. A combined deep-learning and deformable-model approach to fully automatic segmentation of the left ventricle in cardiac MRI. Med Image Anal 2016;30:108-19. [Crossref] [PubMed]
- Cheng JZ, Ni D, Chou YH, et al. Computer-Aided Diagnosis with Deep Learning Architecture: Applications to Breast Lesions in US Images and Pulmonary Nodules in CT Scans. Sci Rep 2016;6:24454. [Crossref] [PubMed]
- Li Z, Wang Y, Yu J, et al. Deep Learning based Radiomics (DLR) and its usage in noninvasive IDH1 prediction for low grade glioma. Sci Rep 2017;7:5467. [Crossref] [PubMed]
- Jia Y, Shelhamer E, Donahue J, et al. Caffe: Convolutional Architecture for Fast Feature Embedding. Available online: https://arxiv.org/pdf/1408.5093.pdf
Convolutional Architecture for Fast Feature Embedding - Skymind Deep Learning 4J, 2017. Available online: https://skymind.ai/
- Abadi M, Agarwal A, Barham P, et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Systems 2015. Available online: https://www.tensorflow.org/
- Wolfram math Wolfram Alpha, 2018. Available online: https://www.wolfram.com/mathematica/
- Al-Rfou R, Alain G, Almahairi A, et al. Theano: A Python framework for fast computation of mathematical expressions. Available online: http://arxiv.org/abs/1605.02688
- Torch, 2017. Available online: http://torch.ch/
- Chollet F. Keras 2018. Available online: https://keras.io/
Neon Nervance Systems 2018 . Available online: https://github.com/NervanaSystems/neonDeep Learning Toolbox Mathworks MATLAB 2018 . Available online: https://au.mathworks.com/help/nnet/ug/deep-learning-in-matlab.html- Oborn BM. Future of medical physics: Real-time MRI-guided proton therapy. Med Phys 2017;44:e77-90. [Crossref] [PubMed]
- Metcalfe P, Alnaghy SJ, Newall M, et al. Introducing dynamic dosimaging: potential applications for MRI-linac. Available online: http://stacks.iop.org/1742-6596/777/i=1/a=012007
- Scrivener M, de Jong EE, van Timmeren JE, et al. Radiomics applied to lung cancer: a review. Transl Cancer Res 2016;5:398-409. [Crossref]
- Suzuki K. Overview of deep learning in medical imaging. Radiol Phys Technol 2017;10:257-73. [Crossref] [PubMed]


