
Diagnostic and prognostic value of KRAS mutations in circulating pancreatic ductal adenocarcinoma tumor DNA
Introduction
Pancreatic cancer is the fourth leading cause of cancer-related death and one of the most malignant tumors (1). Pancreatic ductal adenocarcinoma (PDAC), the most common type of pancreatic tumor, accounts for about 85% of cases (2). More than 25,000 patients are diagnosed with PDAC every year, and a significant portion of them die. The 5-year survival rate for PDAC patients is less than 10% (3). Thus far, surgical resection is the only viable and curative treatment for PDAC. However, less than 20% of PDAC patients are able to have curative surgery, with the majority of them presenting at a very late stage during diagnosis. This may be because of the lack of an effective method for early detection (4-6). Several tumor biomarkers, such as carbohydrate antigen (CA) 19-9 and carcinoembryonic antigen (CEA), have been used in clinical work for the diagnosis and prognosis prediction of PDAC patients. Although they are advantageously of low cost, simplistic, and minimally invasive, their sensitivity and specificity are not sufficient to achieve effective early diagnosis (7,8). Therefore, new effective biomarkers are urgently needed for diagnosis, disease monitoring, and treatment selection for PDAC patients.
The cell-free component of peripheral blood (circulating tumor DNA, ctDNA) can be found in the plasma of patients with cancer and has been found to contain gene mutations representative of primary tumors (9). Plasma ctDNA mutations have significant diagnostic and prognostic value for cancer patients (10). In addition, since plasma ctDNA samples can be repeatedly and non-invasively obtained, they can be used to monitor the response of cancer patients in real time during treatment or disease progression (11,12). Digital polymerase chain reaction (PCR) has been applied to analyze plasma ctDNA for specific gene mutations in several cancers such as colorectal cancer (13-16). However, few studies have been performed analyzing the diagnostic and prognostic value of plasma ctDNA in patients with PDAC before and after surgery.
Mutations in the Kirsten rat sarcoma vial oncogene homolog (KRAS) gene are present in the majority of PDAC cases (17,18). The reported KRAS mutation rate ranges from 75% to 95% (19). The KRAS gene in PDAC is an oncogene that is constitutively active by a point mutation in codon 12 (20), which is thought to occur early in the tumor. Some KRAS mutations may have an impact on the survival of the patients (21,22).
In this study, we not only compared preoperative plasma ctDNA (pre-ctDNA) with surgical tissue DNA (tDNA), but also analyzed plasma ctDNA KRAS mutations in resectable PDAC patients before and after surgery. We evaluated the diagnostic, treatment, and prognostic value of pre-ctDNA, postoperative plasma ctDNA (post-ctDNA), and the changes in pre-ctDNA and post-ctDNA abundance (pre-post ctDNA) in resectable PDAC.
Methods
Patient characteristics
Surgical specimens and plasma samples were obtained from a total of 73 patients with pancreatic lesions at the Peking University Cancer Hospital between June 2016 and May 2017. Among these patients, 35 cases were reported as PDAC by postoperative pathology and were involved in our study. Thirty-eight cases reported as either pancreatic neuroendocrine neoplasm, duodenal papillary carcinoma, or benign pancreatic tumors were eliminated. All 35 PDAC patients were Chinese and agreed to pancreaticoduodenectomy or total pancreaticoduodenectomy through the Department of Hepato-Pancreato-Biliary Surgery, Peking University Cancer Hospital. The pathologic stage of residual tumours (R) was determined according to the eighth edition of the AJCC cancer staging for pancreatic cancer. R0 resections showed no tumour residues, and R1 resections showed microscopically positive margins (23,24). All 35 patients with PDAC in our study received R0 resection, but not R1 resection. And patients did not receive anticancer treatment before the operation. The details of these patients are listed in Table 1. This study was approved by the Ethics Committee of Beijing Cancer Hospital and carried out in accordance with the approved guidelines. All the patients agreed to the study and signed written informed consent.

Full table
DNA extraction and sanger sequencing
All fresh tissue samples, preserved in liquid nitrogen, were obtained during surgery and not from biopsy. Tissue DNA was extracted from 35 pairs of resectable PDAC patients using EasyPure Genomic DNA Kits (TransGen Biotech, Beijing, China) according to the manufacturer’s instructions. The DNA concentration was determined by NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA). DNA KRAS mutations (G12V, G12D and G12R) were identified by Sanger sequencing. Primer sequences were as follows:
- Forward: 5'-GCAGAACAGCAGTCTGGCTA-3';
- Reverse: 5'-TGGACCCTGACATACTCCCA -3'.
Plasma samples and extraction of ctDNA
A total of 35 pairs of peripheral plasma samples were collected from 35 resectable PDAC patients one day before surgery and ten days after surgery. Blood samples (5 mL) were placed in EDTA-containing tubes and processed within 2 hours after collection. Blood was separated via centrifugation at 1,900 g for 10 min at 4 °C and centrifuged at 16,000 g for 10 min at 4 °C to eliminate cell debris. And ctDNA was isolated from 2 mL of plasma using the QIAamp Circulating Nucleic Acid Kit (Qiagen, QIAGEN Strasse 1, 40724 Hilden, Germany) according to the manufacturer’s instructions. The plasma ctDNA concentration was quantified using a Qubit® 2.0 fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). Plasma ctDNA samples were stored at −80 °C until further use.
Plasma ctDNA KRAS mutations via digital PCR
We used a QuantStudio™ 3D Digital PCR System (Thermo Fisher Scientific, Waltham, MA, USA) to detect plasma ctDNA KRAS Mutations by QuantStudi™ 3D Digital PCR 20K Chip. In brief, reactions were transferred to each labeled reaction tube in 15 µL of reaction volume, which consisted of 5 µL of extracted plasma ctDNA, 7.5 µL of Master Mix v2 (2×), 0.375 µL of TaqMan® Digital PCR Liquid Biopsy Assays, and 2.125 µL of nuclease-free water. The digital PCR cycling protocol was as follows: 96 °C for 10 minutes, 39 cycles at 60 °C for 2 minutes and 98 °C for 30 seconds, 60 °C for 2 minutes, and a final step at 10 °C to infinity. QuantStudio™ 3D AnalysisSuite™ Software was used to analyze the data.
In this study, we performed digital PCR on the three most common KRAS mutations in codon 12 (G12V, G12D, and G12R) because these three types of KRAS mutations encompass nearly all KRAS mutations in pancreatic cancer (25). And our tissue samples revealed that only these three types of KRAS mutations were detected by Sanger sequencing.
- The digital PCR probe sequences were as follows:
- G12V(KRAS_520,VIC(wild-type)/FAM(mutant-type): CGTCAAGGCACTCTTGCCTACGCCA[C/A]CAGCTCCAACTACCACAAGTTTATA);
- G12D(KRAS_521,VIC/FAM: CGTCAAGGCACTCTTGCCTACGCCA[C/T]CAGCTCCAACTACCACAAGTTTATA);
- G12R(KRAS_518,VIC/FAM: GTCAAGGCACTCTTGCCTACGCCAC[C/G]AGCTCCAACTACCACAAGTTTATAT).
Statistical analysis
In our data analysis, the abundance of KRAS mutations is defined by FAM/(FAM + VIC). The pre-post ctDNA is defined by pre-ctDNA abundance − post-ctDNA abundance. Therefore, a positive value from this calculation represents a decrease in the number of KRAS mutations after surgery (i.e., pre-post ctDNA −). A negative value for the difference in pre-ctDNA and post-ctDNA indicates an increase in the number of KRAS mutations after surgery (i.e., pre-post ctDNA +).
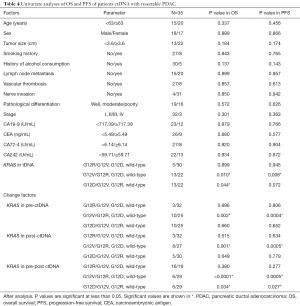
Patients with resectable PDAC were subjected to statistical analyses based on the following variables: age, sex, tumor size, smoking history, history of alcohol consumption, lymph node metastasis, vascular thrombosis, nerve invasion, pathological differentiation, stage, CA19-9, CEA, CA72-4, CA242, tDNA (G12R, G12V or G12D), pre-ctDNA (G12R, G12V or G12D), post-ctDNA (G12R, G12V or G12D), and pre-post ctDNA (G12R, G12V or G12D). Statistical analysis was performed using Kappa and McNemar’s tests to analyze the consistency of KRAS mutations between surgical tDNA and pre-ctDNA. Statistical analysis was performed using Chi-square test and Fisher’s Exact Test to analyze the correlations between pre-ctDNA or post-ctDNA and clinicopathological parameters.
All 35 patients were evaluated for overall survival (OS) and progression-free survival (PFS). OS was defined as the elapsed time between the inclusion date and death due to any cause. PFS was defined as the elapsed time between the date of inclusion and the date of tumor progression. All 35 patients did not die within 30 days after surgery. After a median follow-up time of 12.4 months (range, 6.1–17.2 months), 8 (22.9%) patients died. The Kaplan-Meier method was used to analyze OS or PFS, and log-rank test was used to estimate the difference. On the basis of the log-rank test, variables with P values less than 0.05 were maintained in the multivariate Cox proportional hazards regression model. All statistical analyses were performed using IBM SPSS statistics version 20. Missing data were automatically excluded from the analyses.
Results
Consistency of KRAS mutations between tDNA and pre-ctDNA
KRAS mutations were detected in 31 of 35 tDNA samples (88.6%). The frequencies of the G12R, G12V, and G12D mutations and wild-type KRAS alleles were 5 of 35 (14.3%), 13 of 35 (37.1%), 13 of 35 (37.1%), and 4 of 35 samples (11.4%), respectively.
The two-dimensional histogram of the digital PCR assay for KRAS amplification is shown in Figure 1. KRAS mutations in pre-ctDNA were seen in 23 of 35 samples (65.7%). The frequencies of the G12R, G12V, and G12D mutations and wild-type KRAS alleles were 3 of 35 (8.57%), 10 of 35 (28.6%), 10 of 35 (28.6%), and 12 of 35 samples (34.3%), respectively. KRAS mutations were consistent between tDNA and pre-ctDNA in 27 of 35 samples (77.1%, kappa index =0.397, P=0.003; Table 2).
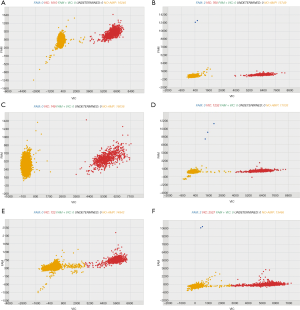

Full table
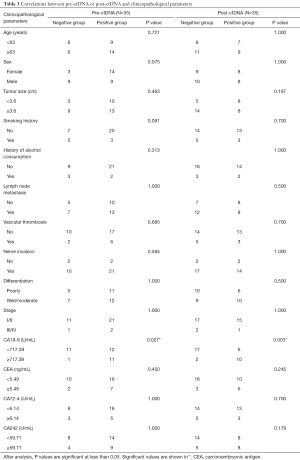
Correlations between pre-ctDNA or post-ctDNA and clinicopathological parameters
Pre- and post-ctDNA showed statistically significant associations with CA19-9 levels before surgery (P=0.027 and P=0.003, respectively). However, no significant correlations between pre- and post-ctDNA and age, sex, tumor size, smoking history, history of alcohol consumption, lymph node metastasis, vascular thrombosis, nerve invasion, pathological differentiation, stage, CEA, CA72-4, and CA242 were found (Table 3).
Full table
Univariate and multivariate analyses of the association between pre-, post-, and pre-post ctDNA and overall survival and progression-free survival
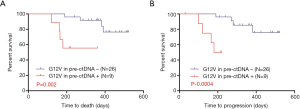
Univariate analyses of pre-ctDNA demonstrated that G12V in tDNA, G12D in tDNA, and G12V in pre-ctDNA showed significant associations with OS (P=0.010, P=0.044, and P=0.002, respectively). G12V in tDNA and G12V in pre-ctDNA were significant predictors of PFS (P=0.008 and P=0.0004, respectively). Multivariate analyses demonstrated that G12V in pre-ctDNA might be an independent predictor of PFS and OS. The OS and PFS curves for G12V in pre-ctDNA are shown in Figure 2.
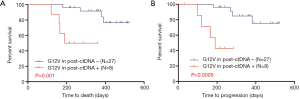
For post-ctDNA, the univariate analyses demonstrated that G12V in tDNA, G12D in tDNA, and G12V in post-ctDNA were significant predictors of OS (P=0.010, P=0.044, and P=0.001, respectively). Moreover, G12V in tDNA and G12V in post-ctDNA (P=0.008 and P=0.0005, respectively) showed significant associations with PFS. Multivariate analyses revealed that G12V in post-ctDNA might be an independent predictor of PFS and OS. The OS and PFS curves for G12V in post-ctDNA are shown in Figure 3.
For pre-post ctDNA, the univariate analyses demonstrated that G12V in tDNA, G12D in tDNA, G12V in pre-post ctDNA, and G12D in pre-post ctDNA were significant predictors of OS (P=0.010, P=0.044, P<0.0001, and P=0.034, respectively). Moreover, G12V in tDNA, G12V in pre-post ctDNA, and G12D in pre-post ctDNA were significant factors associated with PFS (P=0.008, P=0.0005, and P=0.027, respectively). Multivariate analyses showed that G12V or G12D in pre-post ctDNA might be independent predictors of OS and PFS. The OS and PFS curves for G12V and G12D in pre-post ctDNA are shown in Figure 4.
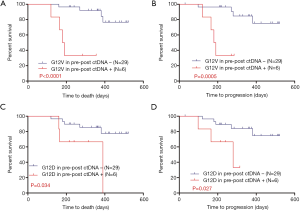
The results of the univariate and multivariate analyses of the association between OS and PFS in patients with resectable PDAC and ctDNA are listed in Tables 4,5.
Full table

Full table
Discussion
In the current study, we found that ctDNA detection in PDAC and its expression level dynamics can be used to determine prognosis and monitor disease (26). Significant progress in the comparative analysis of KRAS gene expression in plasma and tissue in pancreatic cancer has been made. Kinugasa et al. reported that the KRAS mutation rates in endoscopic ultrasound-guided fine-needle aspiration biopsy tDNA and pre-ctDNA were 74.7% and 62.6%, respectively, with a consistency rate of 58 of 75 samples (77.3%) (27). In our study, the KRAS mutation rates in tDNA and pre-ctDNA were 88.6% and 65.7%, respectively, and the consistency rate of KRAS mutations between tDNA and pre-ctDNA was 27 of 35 samples (77.1%, kappa index =0.397, P=0.003). Our findings indicate good but not high consistency between tDNA and pre-ctDNA. This may be due to the small sample size of this study. Therefore, a future study with a larger sample size is needed to verify consistency. Another conceivable reason may be the limitations of ctDNA detection. If the tumor is not affected by other factors such as tumor progression or metastasis, ctDNA may not be released into the blood and cannot be detected.
Currently, serum protein biomarker levels such as CA19-9 are used to monitor and evaluate the treatment of patients with PDAC; CA19-9 levels before surgery are inversely associated with survival. In our study, we found significant correlations between pre-ctDNA, post-ctDNA, and CA19-9 levels before surgery. The results of our analyses suggest that the higher the CA19-9 level before surgery, the higher the positive rate of ctDNA before or after surgery. Therefore, in patients with PDAC, the dynamic monitoring of ctDNA before and after surgery and CA19-9 levels before surgery are complementary and have a good clinical diagnostic value.
Previous research has indicated that the reason the postoperative levels of ctDNA in PDAC are lower than those of preoperative ctDNA may be a significant reduction in tumor burden. Moreover, the increase in postoperative ctDNA may be due to ctDNA release caused by tissue injury during surgery. Another conceivable reason for the increase in postoperative ctDNA levels may be recurrence or tumor metastasis (28). Importantly, most studies on ctDNA in PDAC only reported on the correlation between pre-ctDNA and OS, lacking post-ctDNA for comparison. Therefore, we focused on the dynamic changes in ctDNA before and after surgery to allow for such a comparison. The dynamic observation of ctDNA can provide more information on the effects of treatment and the treatment strategy in patients with PDAC. Our study suggests that dynamic changes in ctDNA before and after surgery may be a very effective and sensitive indicator for PDAC.
Hadano et al. reported that, among 105 PDAC patients who underwent pancreatoduodenectomy, the length of OS was 13.6 months in patients with ctDNA mutations and 27.6 months in those without ctDNA mutations. Patients with ctDNA mutations had a significantly poorer prognosis with respect to OS (29). Our study found that the G12V mutation in pre-ctDNA, post-ctDNA, and pre-post ctDNA was associated with OS or PFS, and might be an independent prognostic factor of OS and PFS. Therefore, patients with resectable PDAC with the ctDNA G12V mutation in pre-ctDNA or post-ctDNA may have a poorer prognosis. Our results also showed that patients with PDAC with an increase in the abundance of pre-ctDNA and post-ctDNA (G12V or G12D) may have a worse prognosis than patients with a decrease in ctDNA KRAS mutations (G12V or G12D). A conceivable reason for this finding may be that the basal GTPase activity of G12V is approximately one-quarter that of G12D and one-tenth that of wild-type KRAS (30-32). Moreover, Rat-1 cells carrying the G12V mutation are more aggressive than wild-type KRAS cells or those with other mutations (33).
There are some limitations to our study. First, the sample size was small. We will enroll a larger number of patients in a future study to provide more evidence for our conclusion. Second, we selected the most common KRAS mutations in this study. More targeted mutations will be included in the future. Third, no negative controls were included in this study. In the future, negative controls will be added to minimize the false positive rate of ctDNA detected by digital PCR. Overall, we hope that the findings of our current study can be confirmed in future studies.
Conclusions
In summary, our research has demonstrated that post-ctDNA targeted KRAS mutations (G12V) and an increase in the abundance of pre-ctDNA and post-ctDNA (G12V or G12D) may be related to disease progression and a worse prognosis for PDAC. Furthermore, our study suggests the G12V KRAS mutation in pre-ctDNA, post-ctDNA, or pre-post ctDNA may be an independent prognostic factor in either univariate or multivariate analyses of OS or PFS. In addition, we also confirm that pre-ctDNA was concordant with tDNA and that pre-ctDNA targeted KRAS mutations (G12V) may be related to disease progression and a worse prognosis for PDAC.
Acknowledgments
Funding: This work was supported by Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding Support (XMLX201708); the Capital Health Research and Development of Special Funds (2016-2-2151); the National Natural Science Funding (31770836, 61571437, 81673500); and the International Science and Technology Cooperation Program of China (2013DFG32720).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.05.33). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was reviewed and approved by the Institutional Review Board of Peking University Cancer Hospital and Institute, Beijing, China (2016KT84). All study participants provided informed written consent prior to study enrollment.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7-30. [Crossref] [PubMed]
- Modolell I, Guarner L, Malagelada JR. Vagaries of clinical presentation of pancreatic and biliary tract cancer. Ann Oncol 1999;10:82-4. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015;65:5-29. [Crossref] [PubMed]
- Kamisawa T, Wood LD, Itoi T, et al. Pancreatic cancer. Lancet 2016;388:73-85. [Crossref] [PubMed]
- Grover S, Syngal S. Hereditary pancreatic cancer. Gastroenterology 2010;139:1076-80, 80 e1-2.
- Yachida S, Jones S, Bozic I, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 2010;467:1114-7. [Crossref] [PubMed]
- Tempero MA, Uchida E, Takasaki H, et al. Relationship of carbohydrate antigen 19-9 and Lewis antigens in pancreatic cancer. Cancer Res 1987;47:5501-3. [PubMed]
- Marrelli D, Caruso S, Pedrazzani C, et al. CA19-9 serum levels in obstructive jaundice: clinical value in benign and malignant conditions. Am J Surg 2009;198:333-9. [Crossref] [PubMed]
- Ma M, Zhu H, Zhang C, et al. "Liquid biopsy"-ctDNA detection with great potential and challenges. Ann Transl Med 2015;3:235. [PubMed]
- Kimura T, Holland WS, Kawaguchi T, et al. Mutant DNA in plasma of lung cancer patients: potential for monitoring response to therapy. Ann N Y Acad Sci 2004;1022:55-60. [Crossref] [PubMed]
- Dawson SJ, Tsui DW, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 2013;368:1199-209. [Crossref] [PubMed]
- Douillard JY, Ostoros G, Cobo M, et al. Gefitinib treatment in EGFR mutated caucasian NSCLC: circulating-free tumor DNA as a surrogate for determination of EGFR status. J Thorac Oncol 2014;9:1345-53. [Crossref] [PubMed]
- Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579-86. [Crossref] [PubMed]
- Tabernero J, Lenz HJ, Siena S, et al. Analysis of circulating DNA and protein biomarkers to predict the clinical activity of regorafenib and assess prognosis in patients with metastatic colorectal cancer: a retrospective, exploratory analysis of the CORRECT trial. Lancet Oncol 2015;16:937-48. [Crossref] [PubMed]
- Reinert T, Scholer LV, Thomsen R, et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut 2016;65:625-34. [Crossref] [PubMed]
- De Mattos-Arruda L, Weigelt B, Cortes J, et al. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Ann Oncol 2014;25:1729-35. [Crossref] [PubMed]
- Bos JL. ras oncogenes in human cancer: a review. Cancer Res 1989;49:4682-9. [PubMed]
- Koorstra JB, Hustinx SR, Offerhaus GJ, et al. Pancreatic carcinogenesis. Pancreatology 2008;8:110-25. [Crossref] [PubMed]
- Almoguera C, Shibata D, Forrester K, et al. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell 1988;53:549-54. [Crossref] [PubMed]
- Barbacid M. ras genes. Annu Rev Biochem 1987;56:779-827. [Crossref] [PubMed]
- Hruban RH, Goggins M, Parsons J, et al. Progression model for pancreatic cancer. Clin Cancer Res 2000;6:2969-72. [PubMed]
- Ogura T, Yamao K, Hara K, et al. Prognostic value of K-ras mutation status and subtypes in endoscopic ultrasound-guided fine-needle aspiration specimens from patients with unresectable pancreatic cancer. J Gastroenterol 2013;48:640-6. [Crossref] [PubMed]
- Demir IE, Jager C, Schlitter AM, et al. R0 versus R1 resection matters after pancreaticoduodenectomy, and less after distal or total pancreatectomy for pancreatic cancer. Ann Surg 2017; [Epub ahead of print]. [Crossref] [PubMed]
- Kamarajah SK, Burns WR, Frankel TL, et al. Validation of the American Joint Commission on Cancer (AJCC) 8th Edition Staging System for Patients with Pancreatic Adenocarcinoma: A Surveillance, Epidemiology and End Results (SEER) Analysis. Ann Surg Oncol 2017;24:2023-30.
- Grünewald K, Lyons J, Frohlich A, et al. High frequency of Ki-ras codon 12 mutations in pancreatic adenocarcinomas. Int J Cancer 1989;43:1037-41. [Crossref] [PubMed]
- Perets R, Greenberg O, Shentzer T, et al. Mutant KRAS Circulating Tumor DNA Is an Accurate Tool for Pancreatic Cancer Monitoring. Oncologist 2018;23:566-72. [Crossref] [PubMed]
- Kinugasa H, Nouso K, Miyahara K, et al. Detection of K-ras gene mutation by liquid biopsy in patients with pancreatic cancer. Cancer 2015;121:2271-80. [Crossref] [PubMed]
- Nakano Y, Kitago M, Matsuda S, et al. KRAS mutations in cell-free DNA from preoperative and postoperative sera as a pancreatic cancer marker: a retrospective study. Br J Cancer 2018;118:662-9. [Crossref] [PubMed]
- Hadano N, Murakami Y, Uemura K, et al. Prognostic value of circulating tumour DNA in patients undergoing curative resection for pancreatic cancer. Br J Cancer 2016;115:59-65. [Crossref] [PubMed]
- Tada M, Omata M, Ohto M. Clinical application of ras gene mutation for diagnosis of pancreatic adenocarcinoma. Gastroenterology 1991;100:233-8. [Crossref] [PubMed]
- John J, Frech M, Wittinghofer A. Biochemical properties of Ha-ras encoded p21 mutants and mechanism of the autophosphorylation reaction. J Biol Chem 1988;263:11792-9. [PubMed]
- Krengel U, Schlichting I, Scherer A, et al. Three-dimensional structures of H-ras p21 mutants: molecular basis for their inability to function as signal switch molecules. Cell 1990;62:539-48. [Crossref] [PubMed]
- Kaplan PL, Ozanne B. Cellular responsiveness to growth factors correlates with a cell's ability to express the transformed phenotype. Cell 1983;33:931-8. [Crossref] [PubMed]


