
Identification of unique microRNAs associated with pancreatic cancer radioresistance using deep sequencing
Introduction
Ductal adenocarcinoma of the pancreas is one of the lethal primary gastrointestinal cancer. In the United States, pancreatic cancer has become the most common cause of cancer-related deaths, and most patients die within 12 months of diagnosis (1). Pancreatic cancer is also the third leading cause of cancer-related deaths in the European Union (2) and by 2030 will be the second leading cause of cancer-related deaths in both genders in the United States (3). A major reason for the poor overall survival is that most patients present with advanced, inoperable disease. For patients with locally advanced pancreatic cancer, the current standard therapeutic protocol consists of gemcitabine-based chemoradiotherapy (4). Unfortunately, pancreatic cancer is often chemo-radioresistant, so this therapy often fails (5). Additional effort is needed to identify biomarkers that could predict the therapeutic response.
MicroRNAs (miRNAs) are small, approximately 18–25 nucleotides (nt) long, non-protein-coding single-stranded RNAs. miRNAs regulate multiple cellular processes, either physiological or pathological, through their target genes, such as apoptosis, proliferation, cell cycle, angiogenesis, and differentiation (6,7). A number of miRNAs have also been shown to be regulated in various cancers and some have been associated with clinical outcomes and radioresistance (8,9). However, involvement of miRNAs in the molecular processes of the cell response to ionizing radiation (IR) in pancreatic cancer remains poorly understood. Therefore, we established a radioresistant pancreatic cancer cell line and examined the miRNA expression differences between the parent pancreatic and the radioresistant cell lines using deep sequencing to identify miRNAs associated with resistance to IR in pancreatic cancer.
Methods
Cells
Pancreatic cancer parental cell lines (SW1990 and PANC-1), as well as the radioresistant equivalent cell lines (SW1990-R and PANC-1-R), were cultured in RPMI-1640 media supplemented with 10% fetal bovine serum (FBS). Cells were cultured in a humidified atmosphere of 5% CO2 at 37 °C.
Irradiation
Parental cells were first grown to approximately 60% confluence and irradiated with 2 Gy (6MV X-radiation, 200 cGy/min). When cells reached approximately 90% confluence, they were subcultured into new flasks and were treated with 2 Gy again when they reached 60% confluence. This procedure was repeated until the total dose was 30 Gy, at which point the cells were irradiated by the same process but with a dose of 5 Gy instead of 2 Gy. This procedure was repeated until the total dose of IR was 60 Gy. Then, cells were irradiated following the same protocol but with a dose of 10 Gy until the total dose of IR was 80 Gy. Multiple irradiated cells are referred to as radioresistant cells.
Colony formation assays
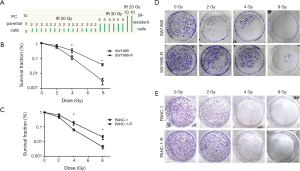
Cells were seeded at a density of 1×106 cells/T25 flask in complete medium. After 24 h, the cells were seeded at a density of 5,000 cells per well in a six-well plate and treated with a single dose of radiation (0, 2, 4, or 8 Gy). The cells were then seeded into new six-well tissue culture plates and incubated for 10 days without changing the culture medium. The cultures were fixed with methanol and stained with crystal violet. The number of colonies with >50 cells was counted under a dissecting microscope. The survival fractions (SFs) were calculated using the following equation: SF = colonies counted/cells seeded × (plating efficiency/100), which takes the individual plating efficiency into consideration. A survival curve was derived using the following multi-target single-hit model: SF =1– (1– e−D/D0)N.
Cell cycle analysis
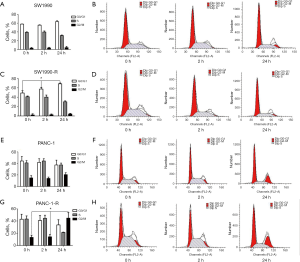
To synchronize the cell cultures, parental cells and radioresistant cells were separately seeded in a six-well plate in growth medium with 10% FBS and grown overnight. Then, the cultures were rinsed with phosphate-buffered saline (PBS) and transferred to serum free medium. Following serum starvation for 24 h, the cells were passaged and released into the cell cycle with the addition of serum. Parental cells and radioresistant cells were separately irradiated with 0 or 5 Gy and fixed at 0, 2, and 24 h post-radiation, cells were harvested, washed in cold PBS fixed in cold 70% ethanol, and stored overnight at 4 °C. Cells were washed twice with PBS and centrifuged at 850× g. Then, samples were incubated with 100 µg/mL RNase and 50 µg/mL propidium iodide for 30 min in the dark. Cells were then analyzed by flow cytometry.
Small RNA deep sequencing
RNA segments of different sizes were separated by polyacrylamide gel electrophoresis (PAGE) into segments of 18–30 nt stripe and recycled. Adaptor ligation: The connection system was prepared, blended, centrifuged, and connected warmly for a period of time. The reverse transcription-polymerase chain reaction (RT-PCR) system was prepared, and PCR amplification was performed. The purified PCR constructs were recovered by PAGE and the recycled products were dissolved in EB solution. Finally, to complete construction of the library, label was added and the Agilent 2100 Bioanalyzer and ABI StepOnePlus Real-Time PCR System were used to test the quality and production of the prepared library. The prepared libraries were loaded into the patterned nanoarrays and single-end reads of 50 bp were read on the BGISEQ-500 platform (BGI, China; http://www.seq500.com/en/).
Analysis of differentially expressed miRNAs
To identify miRNAs differentially expressed between SW1990 and SW1990-R cells, we applied transcripts per million (TPM) to normalize the expression of miRNAs in the two groups. We calculated the fold change (FC) and P value via t-test, and corrected the P value into a false discovery rate (FDR) using the Benjamin and Hochberg method (10). FDR ≤0.05 and |log2FC| ≥2 were set as the cutoffs to screen differentially expressed miRNAs.
Confirmation of miRNA expression levels
To detect the sequencing results, qRT-PCR was performed as the protocol of All-in-One qPCR Mix (GeneCopoeia, Inc.) described. Total RNA from SW1990 and SW1990-R cells was extracted using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA). All assays were carried out in triplicate. Each sample was normalized based on expression of the housekeeping gene (RNU6B). The specificity of amplification was confirmed by melting curve analysis and also by running PCR products on agarose gel (3%) electrophoresis. Relative quantification of target miRNA expression was evaluated using the comparative cycle threshold (CT) method (11).
Bioinformatics analysis
The 49 nt sequence tags from the BGISEQ-500 sequencing were first subjected to data cleaning analysis, which removes low quality tags and the 5' adaptor contaminants from the 50 nt tags, to obtain credible, clean tags. The length distribution of the clean tags and common and specific sequences between samples was then summarized. Standard analysis annotates the clean tags into different categories and uses those which cannot be annotated to any category to predict novel miRNAs and seed edit of potential known miRNAs. After obtaining the miRNA results, target prediction for miRNAs and GO enrichment and KEGG pathway analyses of the target genes were performed.
Transfection
Radioresistant cells were suspended in RPMI-1640 medium supplemented with 10% FBS, seeded in six-well plates (1×106 cells/well), and transfected with miR-216a mimic or miR-Ctrl (20 µM; GenePharma, Shanghai, China) with Lipofectamine™ RNAiMAX (Invitrogen) according to the manufacturer’s instruction. After 24 h of transfection, cells were treated with 5 Gy of radiation. Then, following 24 h of radiation (48 h of transfection), samples were harvested for Western blotting. For colony formation assays, following 48 h of radiation (72 h of transfection), complexes were removed and change medium, cells were grown for 10 days.
Western blotting
Equal amounts of protein were resolved using sodium dodecyl sulfate PAGE, transferred onto a polyvinylidene fluoride membrane, and blocked with a 5% dry milk at room temperature for 2 h. The membranes were incubated with primary antibodies at 4 °C overnight. After being blotted for 1 hour at room temperature with an appropriate secondary antibody, the membranes were exposed to enhanced chemiluminescence detection reagents (Beyotime, Shanghai, China). Primary antibodies against JAK2 and GAPDH were obtained from Wanleibio (Wanleibio Shenyang, China), anti-STAT3 was obtained from Abcam (Abcam, Cambridge, UK), and anti-pSTAT3 was obtained from Cell Signaling Technology (Beverly, MA, USA).
Statistical analysis
All numerical statistical analyses were performed using SPSS version 19.0 (SPSS Inc., Chicago, IL, USA). The data are presented as mean values ± standard deviation from at least three independent experiments. Statistical analysis was performed by two-tailed Student’s t-test. P values <0.05 were considered statistically significant.
Results
Establishment and determination of pancreatic cancer radioresistant cells
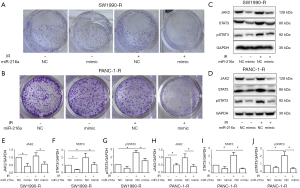
We established radioresistant cell lines derived from the human pancreatic cancer parental cell lines through multiple increasing intensity fractions of IR. A schematic describing the establishment of the radioresistant pancreatic cancer sublines is shown in Figure 1A. Multiply irradiated cells (80 Gy in total) were named SW1990-R AND PANC-1-R. To determine the radiosensitivity of radioresistant cells, a clonogenic survival assay was performed and the fraction of surviving cells was calculated. Figure 1B,C,D,E shows that radioresistant cells demonstrated a significant increase in resistance to IR compared with the parental cell lines. The cell cycle distributions of the parental cells were similar at each time point (0, 2, and 24 h); however, in radioresistant cells, cell cycle analysis revealed the accumulation of cells in G0/G1 or G2/M phase at 24 h post-radiation compared with the earlier time points (Figure 2; P<0.05). The cells that survived multiple fractions of IR were more resistant to IR than the parental cells.
Differentially expressed miRNAs in SW1990 and SW1990-R cells
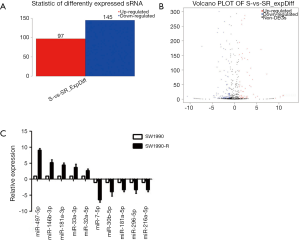
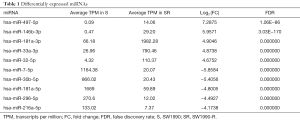
We performed global miRNA expression analysis of radioresistant and parental SW1990 cell lines. In total, 242 significantly differentially expressed miRNAs were identified between the two groups (log2 ratio >1 S/SR). Figure 3A and 3B show that 97 miRNAs were upregulated and 145 were downregulated in radioresistant cells. The ten miRNAs with the greatest relative upregulation or downregulation are shown in Table 1. To confirm the high throughput sequencing results, we performed qRT-PCR to assess the expression of ten miRNAs and the relative expression level of each miRNA is shown in Figure 3C. Expression levels of these ten miRNAs were dramatically different between the radioresistant SW1990-R and parental SW1990 cell lines, consistent with the sequencing data.
Full table
GO and KEGG pathway analyses
The differentially expressed genes were analyzed by GO enrichment and KEGG pathway analysis. GO enrichment analysis groups genes into three categories: molecular function (MF), cellular component (CC), and biological process (BP). The microarray data obtained in the present study revealed that the differentially expressed miRNAs were enriched for GO terms related to the binding and catalytic activity in the cytosol (MF); cell, cell part, and organelle (CC); and cellular process, single-organism process, metabolic process, and biological regulation (BP). The GO terms with more than 3,000 genes in each of the three categories are shown in Figure 4A,B,C. In other words, the function of the target genes of the differentially expressed miRNAs might be involved in cell proliferation, cell cycle, and immunology associated with the radioresistance of pancreatic cancer.
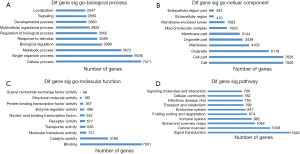
We conducted pathway analysis of the differentially expressed miRNAs using the latest KEGG database. We identified 62 pathways corresponding to differentially expressed miRNAs, and the predominant pathways are shown in Figure 4D. The top ten pathways were signal transduction, cancer: overview, global and overview maps, immune system, folding sorting and degradation, endocrine system, transport and catabolism, infectious disease: viral, cellular community, and signaling molecules and interaction. Thus, the various pathways might affect the radioresistance of pancreatic cancer by targeting differentially expressed these genes.
miR-216a decreased the radioresistance of radioresistant cells by targeting JAK2
To validate whether the differentially expressed miRNAs could affect the cell sensitivity towards radiotherapy, radioresistant cells were treated with radiation following miR-216a NC/mimic transfection. As shown in Figure 5A and 5B, cell growth and proliferation were significantly inhibited following upregulation of miR-216a. To further test whether miR-216a is able to regulate radioresistance by targeting JAK2, miR-216a NC or mimic was transfected into radioresistant cells with or without radiation. Our results showed that radiation dramatically upregulated JAK2 expression in all these groups compared with parallel samples without radiation, whereas the overexpression of miR-216a with miR-216a mimics led to a notable decrease in JAK2/STAT3 signaling pathway protein levels compared with the negative control in radioresistant cells (Figure 5C,D,E,F,G,H,I,J).
Discussion
Radiotherapy is one of the main treatment choices for pancreatic cancer patients. In the present study, we compared differences in miRNA expression in radioresistant and parental stable pancreatic cell lines with RNA sequencing technology. Our aim was to screen and identify miRNAs that might be closely associated with radioresistance of PC cells, as identified miRNAs could be potential therapeutic targets to promote radiosensitivity in PC.
First, we generated radioresistant cell sublines by treating parental cells with a total dose of 80 Gy IR and characterized their radioresistant phenotypes. Radioresistant cells demonstrated a higher potential to form colonies after irradiation when compared with parental cells. Similar results were published by Wang et al. who established radioresistant cell lines derived from the human pancreatic cancer cell lines BxPC3 and PANC-1 by fractionated IR exposure with a total dose of 80 Gy (12). When the cell cycle was analyzed, no difference was apparent in control parental cells (0 Gy) grown for 2 and 24 h after IR treatment. However, the G0/G1 or G2/M phases were found to be longer in radioresistant cells (P<0.05).
To the best of our knowledge, our study was the first to use high-quality sequencing to analyze miRNA expression. Accordingly, high-quality sequence data were obtained for 242 miRNAs with significantly different levels of expression, with 97 upregulated and 145 downregulated miRNAs in radioresistant cells. We confirmed the upregulated and downregulated miRNAs with qRT-PCR, which included miR-497-5p, miR-146b-3p, miR-181a-3p, miR-33a-3p, and miR-32-5p (upregulated); and miR-7-5p, miR-30b-5p, miR-181a-5p, miR-296-5p, and miR-216a-5p (downregulated). Several previous studies have observed that these miRNAs are involved in tumor processes. Zhu et al. observed that the expression of miR-497 was markedly upregulated in temozolomide (TMZ)-resistant glioma cells; high miR-497 expression was associated with the TMZ-resistant phenotype of glioma cells (13). Jafarzadeh et al. demonstrated that, similar to our study, overexpression of hsa-miR-497-5p in HEK293t cells resulted in cell cycle arrest in G0/G1 phase, and the targeting of SMAd3 may regulate the transforming growth factor (TGF) β signaling pathway (14). Riesco-Eizaguirre et al. showed that miR-146b-3p might be a therapeutic target to modulate thyroid cell differentiation and improve the treatment of advanced thyroid cancer (15). In addition, miR-32-5p is associated with cancer-specific survival of patients with high grade, non-muscle-invasive bladder tumors (16). Many studies have observed that miR-7 is downregulated in digestive system malignancies compared with normal tissue (17), which is consistent with our observation that miR-7 is significantly downregulated in radioresistant cells. There are many reports that miR-7 is related to drug resistance in tumors. Kabir et al. demonstrated that miR-7 effectively silenced TYRO3 expression in sorafenib-sensitive cells in human hepatocellular carcinoma (18). miR-7 was also shown to sensitize gastric cancer cells to cisplatin by targeting mammalian target of rapamycin (mTOR) (19). Collectively, our results demonstrate that miR-7 is a potential therapeutic target for the treatment of human pancreatic cancer. In addition, miR-30b-5p has been shown to be downregulated in renal cancer tissues and cell lines, and to regulate the epithelial-mesenchymal transition (EMT) in renal cancer cells (20). Studies have shown that miR-216a enhances the radiosensitivity of pancreatic cancer cells (21). Finally, miR-296-5p was shown to be involved in resistance to radiotherapy and tumor recurrence in early-stage laryngeal carcinoma (22). Consistent with our study, downregulation of miR-296-5p levels was detected in both non-small cell lung cancer tissues and cell lines (23). In addition, miR-296-5p has been linked with glioblastoma invasiveness (24). More promisingly, miR-296-5p inhibited transcriptional mechanisms that support glioblastomas stem cells (25).
It is known that miRNAs function not through one single gene, but through a large network that includes many genes. The biological functions and signaling pathways were associated with the miRNAs identified in the present study, including GO and KEGG pathway enrichment analysis. The GO enrichment analysis found that the target genes of these dysregulated miRNAs were associated with binding (MF), cell (CCs), and cellular process (BPs). In addition, many of the GO terms identified in our results have been reported in other cancer radioresistance studies. Su et al. observed that the cellular response is of great importance during the development of radioresistant esophageal cancer cells (26). Positive regulation of transcription, intracellular protein transport, proteolysis, and cell cycle arrest have been reported to be involved in the radioresistance of non-small cell lung cancer (27). This suggests that the miRNAs identified in this study may regulate radioresistance of pancreatic cancer cells by influencing the expression of these GO database genes. Pathway analysis revealed a total of 42 pathways corresponding to the differentially expressed miRNAs identified. Some of the top ten pathways identified have previously been associated with radioresistance. The PAK4/PPARγ/Nox1 signaling pathway has been reported as a key mediator of tumor cell responsiveness to radiation (28). This PAK4/PPARγ pathway accelerates the EMT in glioma, and consequently, its activation leads to therapy resistance (28). Ciccarelli et al. found that the MEK/ERK pathway plays a prominent role in maintaining the stem-like phenotype of rhabdomyosarcoma cells as well as their innate radioresistance (29). Phosphatidylinositol signaling has been reported to be a key mediator of tumor cell responsiveness to radiation. The phosphatidylinositol 3-kinase (PI3K)/Akt pathway accelerates the repair of DNA following exposure to IR, and consequently, its activation combines with radiotherapy resistance (30). Agliano et al. also reported that PI3K/mTOR inhibitors were useful in radiotherapy resistance in glioblastoma patients (31). Maachani et al. found that FOXM1, which is associated with cell cycle progression and DNA repair, regulated the STAT3 pathway, which contributes to radioresistance in glioblastoma (32). The differentially expressed miRNAs identified in this study may regulate radioresistance in pancreatic adenocarcinoma through these classical pathways.
This study found that radioresistant pancreatic cancer cells could increase the radiosensitivity through upregulated expression of miRNA-216a, furthermore, miRNA-216a may be increase the radiosensitivity of pancreatic cancer cells by inhibiting the JAK2/STAT3 signaling pathway. Our previous study showed that miR-216a negatively regulated the development of pancreatic cancer cells by targeting JAK2 (33). Zhang et al. also confirmed that miRNA-216a may be increase the radiosensitivity of pancreatic cancer cells differently, inhibiting irradiation-induced autophagy by regulating Beclin-1 (21). However, further studies are required to determine whether miR-216a could serve as a potential diagnostic and therapeutic target in pancreatic cancer.
Conclusions
However, this study had some limitations. One limitation is the small sample size; accordingly, it will be necessary to validate the experiments with a larger sample size. Additionally, sequencing was performed on a single microarray platform. Based on the evidence presented above, we have reached the general conclusion that numerous differentially expressed miRNAs may play an important role in radioresistance, and our data suggests that various miRNAs may be predictive biomarkers or therapeutic targets in pancreatic cancer.
Acknowledgments
Funding: This work was financially supported by funding from the Natural Science Foundation of Liaoning Province (No. 201602863).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.06.02). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7-30. [Crossref] [PubMed]
- Ferlay J, Partensky C, Bray F. More deaths from pancreatic cancer than breast cancer in the EU by 2017. Acta Oncol 2016;55:1158-60. [Crossref] [PubMed]
- Rahib L, Smith BD, Aizenberg R, et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 2014;74:2913-21. [Crossref] [PubMed]
- Burris HA, Moore MJ, Andersen J, et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J Clin Oncol 1997;15:2403-13. [Crossref] [PubMed]
- Hidalgo M. Pancreatic cancer. N Engl J Med 2010;362:1605-17. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [Crossref] [PubMed]
- Farazi TA, Spitzer JI, Morozov P, et al. miRNAs in human cancer. J Pathol 2011;223:102-15. [Crossref] [PubMed]
- Zhang Y, Zheng L, Ding Y, et al. MiR-20a Induces Cell Radioresistance by Activating the PTEN/PI3K/Akt Signaling Pathway in Hepatocellular Carcinoma. Int J Radiat Oncol Biol Phys 2015;92:1132-40. [Crossref] [PubMed]
- Korpela E, Vesprini D, Liu SK. MicroRNA in radiotherapy: miRage or miRador? Br J Cancer 2015;112:777-82. [Crossref] [PubMed]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 1995;57:289-300.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Wang P, Zhang J, Zhang L, et al. MicroRNA 23b regulates autophagy associated with radioresistance of pancreatic cancer cells. Gastroenterology 2013;145:1133-43.e12. [Crossref] [PubMed]
- Zhu D, Tu M, Zeng B, et al. Up-regulation of miR-497 confers resistance to temozolomide in human glioma cells by targeting mTOR/Bcl-2. Cancer Med 2017;6:452-62. [Crossref] [PubMed]
- Jafarzadeh M, Soltani BM, Dokanehiifard S, et al. Experimental evidences for hsa-miR-497-5p as a negative regulator of SMAD3 gene expression. Gene 2016;586:216-21. [Crossref] [PubMed]
- Riesco-Eizaguirre G, Wert-Lamas L, Perales-Paton J, et al. The miR-146b-3p/PAX8/NIS Regulatory Circuit Modulates the Differentiation Phenotype and Function of Thyroid Cells during Carcinogenesis. Cancer Res 2015;75:4119-30. [Crossref] [PubMed]
- Lenherr SM, Tsai S, Silva Neto B, et al. MicroRNA Expression Profile Identifies High Grade, Non-Muscle-Invasive Bladder Tumors at Elevated Risk to Progress to an Invasive Phenotype. Genes-Basel 2017;8:77. [Crossref] [PubMed]
- Chen WQ, Hu L, Chen GX, et al. Role of microRNA-7 in digestive system malignancy. World J Gastrointest Oncol 2016;8:121-27. [Crossref] [PubMed]
- Kabir TD, Ganda C, Brown RM, et al. A microRNA-7/growth arrest specific 6/TYRO3 axis regulates the growth and invasiveness of sorafenib-resistant cells in human hepatocellular carcinoma. Hepatology 2018;67:216-31. [Crossref] [PubMed]
- Xu N, Lian YJ, Dai X, et al. miR-7 Increases Cisplatin Sensitivity of Gastric Cancer Cells Through Suppressing mTOR. Technol Cancer Res Treat 2017; [Crossref] [PubMed]
- Liu W, Li H, Wang Y, et al. MiR-30b-5p functions as a tumor suppressor in cell proliferation, metastasis and epithelial-to-mesenchymal transition by targeting G-protein subunit alpha-13 in renal cell carcinoma. Gene 2017;626:275-81. [Crossref] [PubMed]
- Zhang X, Shi H, Lin S, et al. MicroRNA-216a enhances the radiosensitivity of pancreatic cancer cells by inhibiting beclin-1-mediated autophagy. Oncol Rep 2015;34:1557-64. [Crossref] [PubMed]
- Maia D, de Carvalho AC, Horst MA, et al. Expression of miR-296-5p as predictive marker for radiotherapy resistance in early-stage laryngeal carcinoma. J Transl Med 2015;13:262. [Crossref] [PubMed]
- Xu C, Li S, Chen T, et al. miR-296-5p suppresses cell viability by directly targeting PLK1 in non-small cell lung cancer. Oncol Rep 2016;35:497-503. [Crossref] [PubMed]
- Lee H, Shin CH, Kim HR, et al. MicroRNA-296-5p Promotes Invasiveness through Downregulation of Nerve Growth Factor Receptor and Caspase-8. Mol Cells 2017;40:254-61. [Crossref] [PubMed]
- Lopez-Bertoni H, Lal B, Michelson N, et al. Epigenetic modulation of a miR-296-5p:HMGA1 axis regulates Sox2 expression and glioblastoma stem cells. Oncogene 2016;35:4903-13. [Crossref] [PubMed]
- Su H, Lin F, Deng X, et al. Profiling and bioinformatics analyses reveal differential circular RNA expression in radioresistant esophageal cancer cells. J Transl Med 2016;14:225. [Crossref] [PubMed]
- Chen X, Xu Y, Liao X, et al. Plasma miRNAs in predicting radiosensitivity in non-small cell lung cancer. Tumour Biol 2016;37:11927-36. [Crossref] [PubMed]
- Kesanakurti D, Maddirela D, Banasavadi-Siddegowda YK, et al. A novel interaction of PAK4 with PPARgamma to regulate Nox1 and radiation-induced epithelial-to-mesenchymal transition in glioma. Oncogene 2017;36:5309-20. [Crossref] [PubMed]
- Ciccarelli C, Vulcano F, Milazzo L, et al. Key role of MEK/ERK pathway in sustaining tumorigenicity and in vitro radioresistance of embryonal rhabdomyosarcoma stem-like cell population. Mol Cancer 2016;15:16. [Crossref] [PubMed]
- Toulany M, Rodemann HP. Phosphatidylinositol 3-kinase/Akt signaling as a key mediator of tumor cell responsiveness to radiation. Semin Cancer Biol 2015;35:180-90. [Crossref] [PubMed]
- Agliano A, Balarajah G, Ciobota DM, et al. Pediatric and adult glioblastoma radiosensitization induced by PI3K/mTOR inhibition causes early metabolic alterations detected by nuclear magnetic resonance spectroscopy. Oncotarget 2017;8:47969-83. [Crossref] [PubMed]
- Maachani UB, Shankavaram U, Kramp T, et al. FOXM1 and STAT3 interaction confers radioresistance in glioblastoma cells. Oncotarget 2016;7:77365-77. [Crossref] [PubMed]
- Wang S, Chen X, Tang M. MicroRNA-216a inhibits pancreatic cancer by directly targeting Janus kinase 2. Oncol Rep 2014;32:2824-30. [Crossref] [PubMed]


