
circRNAs and miRNAs in the diagnosis of hepatocellular carcinoma: the long race to new targets
The work by Yu and colleagues, recently published on the Journal of Hepatology (1) represents the latest step on a greatly debated and cherished argument: the mechanisms and clinical applicability of circular RNAs (circRNAs) and other non-coding RNAs (in primis microRNAs) in human tumoral disease, and in hepatocellular carcinoma (HCC) in particular. From their discovery in plants in 1976 no particular attention was attributed to the role of circRNAs in human, as they were considered as viroids (2). However, since 2012 we are experiencing a fast-growing interest towards circRNA functions in human: respectively 3 articles were published in 2013, 37 in 2015, 241 in 2017 and already 168 articles are available in May 2018 (source: https://www.ncbi.nlm.nih.gov/pubmed). circRNAs are very stable RNA structures, closed by a continuous loop, present in cell cytoplasm (3). They are highly represented in eukaryotic cells: up to now thousands of circRNAs have been identified in human. Among the circRNA functions, to “sponge” miRNAs is the most important: in this way circRNAs regulate miRNA concentration and—as a consequence—regulate the expression of several genes at transcriptional or post-transcriptional level (4).
In the very elegant study by Yu et al., which included in vitro assays as well as animal and human models, the initial RNA-seq analysis performed on 5 HCC and 5 adjacent non-tumoral tissues identified 13,686 differentially expressed circRNAs. To identify the main key targets in HCC the authors filtered methodically the 13,686 detected circRNAs. Some of the main steps were the exclusion of the low abundance circRNAs (4,727 remaining) considering only novel circRNA. Then, through the comparison with known genes [2,322] encoding for circRNAs [4,142] and the Ingenuity Pathway Analysis, 236 circRNAs were identified. Based on their expression levels, the authors selected 16 circRNAs to be tested on 40 HCC and 40 control tissues; 10 circRNAs were validated (Table 1). In particular, the authors demonstrated in vitro that cSMARCA5 is an abundant, circular and stable transcript. Down-regulation of cSMARCA in HCC was elucidated throw in vitro and in vivo experiments: indeed, the DHX9 up-regulation in HCC prevents the formation of cSMARCA5. Finally, the downregulation of SMARCA5, studied in a new cohort of 163 patients, was associated with the tumor aggressiveness-growth and metastatic potential- and low survival rate. Low level of cSMARCA5 also induced the downregulation of the tumor-suppressor protein TIMP3; indeed overexpression of cSMARCA5 sponges the microRNAs miR-17-3p and miR-191b-5p (1). One additional point of interest is represented by the fact that miR-17-3p belongs to the miR-17-92 cluster, recently described as important for HCC resistance to systemic therapy with sorafenib (5).
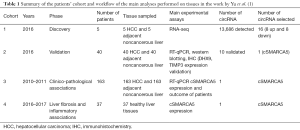
Full table
This represents the last of a long series of recent findings about the association of circRNA-miRNA-mRNA and human neoplasms, as HCC (Figure 1). It was recently described how the hsa_circ_0003570 progressively decreased from normal liver to cirrhosis to HCC, with the lowest concentrations in high-grade HCCs with bigger dimensions (6). Most papers demonstrated that these circRNAs sponge at least one oncogenic or tumor-suppressor miRNA, also helping in the knowledge of miRNAs and other non-coding RNAs in the human tumor biology. For example, the up-regulation of hsa_circ_0000673 promotes the sponging of miR-767-3p, bringing to a SET-mediated increase in HCC cell proliferation and invasion capability (7). Cdr1as showed oncogenic functions in HCC, by sponging miR-7 (8), hsa_circ_0005075 correlated with HCC tumor size, and sponged different miRNAs (3). Another study, in vitro and with animal model, found that circHIPK3 positively regulated the AQP3 gene via miR-124 sponging, promoting HCC cell proliferation and migration (9).
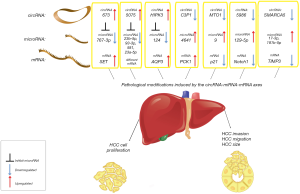
On the other hand, other circRNAs show tumor-suppressor activity: the down-regulation of circC3P1 promotes HCC progression and aggressiveness, decreasing the miR-4641 sponging and activating PCK1 (10). hsa_circRNA_0007874/hsa_circRNA_104135 (or mitochondrial translation optimization 1 homologue, circMTO1) suppresses HCC progression by sponging miR-9 and increasing p21 expression (11). Decreased hsa_circ_0005986 (with consequent increased miR-129-5p) in HCC was correlated with tumor dimensions, microvascular invasion and other pejorative prognostic factors (12).
The roles of circRNA are still largely unclear, and the current studies focus on their interaction with RNAs and protein: the functions of their secondary structures and their roles in DNA genome are not known. In literature, there is a huge growing number of new circRNAs involved in complex pathway interactions, most of them still unknown. However, the quick development of high-throughput RNA deep sequencing, along with the bioinformatics predictor tools, will enable a faster discovery of the pathways interactions of circRNAs (13). Considering their regulator properties, circRNAs might become very important for both diagnosis and therapeutic applications. For example, an in vivo increase in sponge expression is likely to provide an alternative to genetic knockouts of miRNAs. An advantage that cannot be neglected is that circRNA expression is highly tissue specific, quite the opposite to existing protein markers, that have low organ specificity: circRNAs—or the circRNA-miRNA axes—could be suitable diagnostic markers, with higher diagnostic sensitivity than traditional markers (14). circRNAs might become diagnostic biomarkers sooner than we expect, especially in HCC. Recently, a meta-analysis study including 1,752 patients with circRNA expression data from 17 publications showed that the expression of circRNA in HCC tissue was more prone to be detected than other tumor types with higher specificity and diagnostic value (15).
The study of circRNAs and miRNAs in human carcinogenesis is especially crucial in a multistep neoplasm such as HCC, in which most patients suffer from well-known pre-neoplastic conditions (i.e., cirrhosis), and the passage from regenerative and dysplastic nodules to openly neoplastic lesion is well documented by the literature (16). In this setting, any possibility to set up a poorly invasive or not invasive early diagnosis of HCC is likely to change the natural history of cirrhotic patients, from prognosis after surgical resection or local therapy to priority in the transplant waiting list. Many efforts have been made in this direction in order to find serum RNA markers—especially serum miRNAs—for the early diagnosis of HCC. miR-122 alone or in combination with other miRNAs seemed the more promising (17), while other serum miRNAs were proposed in combination with alpha-fetoprotein (AFP) for the early diagnosis of HCC in cirrhotic patients (18). From this point of view, it would be interesting in the future to verify if cSMARCA5 levels in exosomes and blood plasma are also associated to HCC aggressiveness and survival rate as Yu and colleagues had demonstrated on tissue (1), and the same is applicable to any circRNA mentioned above. The possibility to regulate miRNAs such as the above-mentioned miR-17-3p is likely to open more perspective also in the response to sorafenib and other systemic therapies (5).
However, if we consider the clinical application of miRNAs, only a few of them have entered the preclinical and clinical stage. This is due in part to the lack of standardization during analysis (e.g., use of controls tissue, normalization methods) (19). Since today the majority of studies on circRNA mainly used the matched non-tumoral tissue as controls (15), but this changes from case to case. The field is still at his infancy and it would be beneficial to define from the beginning standard regulations for the analysis of circRNAs and miRNAs, especially in a tissue like liver, in which the “normality” itself is difficult to define, due to several changes incurring with age and chronic diseases.
In conclusion, the study of circRNAs and their interactions with miRNAs will be of great help in improving the knowledge about these non-coding RNAs, and the increasing interest of the scientific community on the argument seem to confirm that a panel of non-coding RNAs with clinical use will be available in commerce soon. As a matter of fact, looking at the gargantuan literature on the topic it seems strange that such panels have not been proposed yet for diagnosis and prognosis in human cancers. Apart from the crucial problem of finding standard controls, another limitation to the use of circRNAs as diagnostic tool is that the detection reagents tend to be expensive. However some promising new methods—such as bioluminescence or high-throughput sequencing—used in miRNA detection can be translated to circRNA detection (15). Thus a fast and cheap detection method for circRNA/miRNA would hopefully become a reality in the future, not only for research purposes but also for clinical use.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Chunlin Ou (Cancer Research Institute of Central South University, Changsha, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.06.20). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Yu J, Xu QG, Wang ZG, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol 2018;68:1214-27. [Crossref] [PubMed]
- Sanger HL, Klotz G, Riesner D, et al. Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A 1976;73:3852-6. [Crossref] [PubMed]
- Shang X, Li G, Liu H, et al. Comprehensive Circular RNA Profiling Reveals That hsa_circ_0005075, a New Circular RNA Biomarker, Is Involved in Hepatocellular Carcinoma Development. Medicine (Baltimore) 2016;95:e3811 [Crossref] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. [Crossref] [PubMed]
- Awan FM, Naz A, Obaid A, et al. MicroRNA pharmacogenomics based integrated model of miR-17-92 cluster in sorafenib resistant HCC cells reveals a strategy to forestall drug resistance. Sci Rep 2017;7:11448. [Crossref] [PubMed]
- Fu L, Wu S, Yao T, et al. Decreased expression of hsa_circ_0003570 in hepatocellular carcinoma and its clinical significance. J Clin Lab Anal 2018;32: [Crossref] [PubMed]
- Jiang W, Wen D, Gong L, et al. Circular RNA hsa_circ_0000673 promotes hepatocellular carcinoma malignance by decreasing miR-767-3p targeting SET. Biochem Biophys Res Commun 2018;500:211-6. [Crossref] [PubMed]
- Yu L, Gong X, Sun L, et al. The Circular RNA Cdr1as Act as an Oncogene in Hepatocellular Carcinoma through Targeting miR-7 Expression. PLoS One 2016;11:e0158347 [Crossref] [PubMed]
- Chen G, Shi Y, Liu M, et al. circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis 2018;9:175. [Crossref] [PubMed]
- Zhong L, Wang Y, Cheng Y, et al. Circular RNA circC3P1 suppresses hepatocellular carcinoma growth and metastasis through miR-4641/PCK1 pathway. Biochem Biophys Res Commun 2018;499:1044-9. [Crossref] [PubMed]
- Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology 2017;66:1151-64. [Crossref] [PubMed]
- Fu L, Chen Q, Yao T, et al. Hsa_circ_0005986 inhibits carcinogenesis by acting as a miR-129-5p sponge and is used as a novel biomarker for hepatocellular carcinoma. Oncotarget 2017;8:43878-88. [PubMed]
- Wang J, Liu K, Liu Y, et al. Evaluating the bias of circRNA predictions from total RNA-Seq data. Oncotarget 2017;8:110914-21. [Crossref] [PubMed]
- Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer 2017;16:94. [Crossref] [PubMed]
- Wang M, Yang Y, Xu J, et al. CircRNAs as biomarkers of cancer: a meta-analysis. BMC Cancer 2018;18:303. [Crossref] [PubMed]
- International Agency for Research on Cancer. WHO classification of tumours of the digestive system. 4th edition. Lyon: IARC, 2010.
- Ding Y, Yan JL, Fang AN, et al. Circulating miRNAs as novel diagnostic biomarkers in hepatocellular carcinoma detection: a meta-analysis based on 24 articles. Oncotarget 2017;8:66402-13. [PubMed]
- Zhang Y, Li T, Qiu Y, et al. Serum microRNA panel for early diagnosis of the onset of hepatocellular carcinoma. Medicine (Baltimore) 2017;96:e5642 [Crossref] [PubMed]
- Fittipaldi S, Vasuri F, Bonora S, et al. miRNA Signature of Hepatocellular Carcinoma Vascularization: How the Controls Can Influence the Signature. Dig Dis Sci 2017;62:2397-407. [Crossref] [PubMed]


