
A novel nanoprobe-based assay for detecting K-ras mutations in plasma and stool samples from patients with pancreatic cancer: value in diagnosis and prognosis evaluation
Introduction
Pancreatic cancer is a kind of highly malignant tumor of the digestive system that has a very poor prognosis, with a 5-year survival rate <5% (1). In recent years, the incidence of pancreatic cancer has exhibited a rising trend. According to the statistics reported by the International Agency for Research on Cancer (2), 3.38 million new cases of pancreatic cancer were diagnosed worldwide in 2012, with a mortality-to-incidence ratio of 0.98. The lack of specific clinical manifestations and reliable tumor biomarkers makes the early diagnosis of pancreatic cancer difficult. As a consequence, more than half of cases are diagnosed at an advanced stage, and only 15–20% have the opportunity for radical surgery (3), which is presently the only cure for pancreatic cancer. Therefore, early diagnosis is extremely important for improving the prognosis of patients with pancreatic cancer, and at present, there is an urgent need to find a reliable screening tool to improve the early diagnosis of this aggressive malignancy.
The development, invasion and metastasis of pancreatic cancer involve the dysregulation of many oncogenes and tumor suppressor genes (4,5), of which K-ras has been the most thoroughly studied due to the nearly ubiquitous presence of K-ras mutations in this malignancy (6,7). Since K-ras mutations are of paramount importance for the initiation of pancreatic cancer (8), the detection of K-ras mutations has been regarded as an important tool for the auxiliary diagnosis and prognostic evaluation of pancreatic cancer (9,10). K-ras mutations can be detected in different types of clinical samples, including pancreatic tissue, pancreatic duct brushings, duodenal aspirates, pancreatic juice, bile, plasma, and stool (9). Invasive techniques for obtaining samples such as pancreatic juice or pancreatic tissue are certainly inappropriate for cancer screening, whereas the non-invasive testing of plasma and stool may be applicable for both the diagnosis of symptomatic patients and early screening, especially in populations (11).
Many methods for detecting K-ras mutations in plasma and stool samples have been developed, such as polymerase chain reaction (PCR)-restriction fragment length polymorphism (12) and peptide nucleic acid clamp PCR (13). However, these techniques are associated with drawbacks of being poorly reproducible, labor-intensive, time-consuming, or expensive, which make these difficult to be used for screening and the early diagnosis of pancreatic cancer (14,15). The advent of nanotechnology has allowed for the development of nanoprobes appropriate for biomolecular measurements (16), and nanoparticles tagged with short DNA segments can be used to detect the genetic sequence in a sample, improving the diagnostic accuracy in biological samples that contain trace amounts of target DNA (17,18). Thus, the combination of nanoprobes for the specific recognition of K-ras mutations with conventional PCR may improve the detection of K-ras mutations in plasma and stool samples obtained from patients with pancreatic cancer.
In the present study, we designed novel fluorescent nanoprobes that target K-ras G12V and G13D mutations, which are the most abundant mutation types in pancreatic cancer (6,7), and determined the diagnostic accuracy of this novel nanoprobe-based assay in the detection of K-ras mutations in plasma and stool samples obtained from patients with pancreatic cancer, subjects with benign pancreatic diseases, and normal controls. Furthermore, we also assessed the correlation of nanoprobe-based plasma and stool K-ras mutation testing with the clinicopathological characteristics and survival of pancreatic cancer patients.
Methods
Patients
A total of 121 patients (64 men and 57 women) treated at the Second Affiliated Hospital of Jiaxing University from June 2013 to October 2015 were included in this study. Among these patients, 58 had pancreatic ductal adenocarcinoma, 18 had chronic pancreatitis, 7 had pancreatic cystadenoma, 5 had pancreatic cyst, and 2 had solid pseudopapillary tumor of the pancreas. All diseases were diagnosed pathologically or by endoscopic ultrasonography-guided fine needle aspiration. In addition, 31 volunteers who underwent a physical examination and exhibited no abnormality were included as normal controls. Informed consent was obtained from all participants, and the study protocol was approved by the Ethics Committee of the Second Affiliated Hospital of Jiaxing University (No. L2015-28).
Sample collection and DNA extraction
Stool specimens were collected, frozen, and stored at −80 °C. After stool (200 mg) was processed with TEN and starch, fecal DNA was prepared using the phenol-chloroform method and purified using a DNA purification kit (QIAGEN, Frankfurt, Germany). Then, peripheral blood samples (5 mL) were taken and centrifuged to collect the supernatant as plasma samples, which were preserved at −80 °C for further use. Plasma DNA was prepared using a magnetic bead DNA extraction kit (Omega, Norcross, USA). The concentration of the prepared DNA was determined using a NanoDrop 1000 spectrophotometer (NanoDrop, Wilmington, DE, USA), and the OD260/OD280 ratio was calculated to ensure that the purity of the DNA was good (OD260/OD280 ratio =1.6–1.9). The prepared DNA was stored at −80°C.
Nanoparticle probe preparation
Magnetic nanoparticles were prepared through the hydrothermal synthesis method. Briefly, 0.01 M of salicylic acid solution was added to a triangle flask in an argon environment. After adjusting the pH value to 11.0 with NaOH solution, Fe3O4 and Fe2O3 were added at a Fe ion concentration ratio of 2:1. After fully mixing and heating to 90 °C for 4 hours, the mixture was centrifuged three times at 4,000 rpm to purify the magnetic nanoparticles. After that, the magnetic nanoparticles were observed using a transmission electron microscope, in order to ensure that these were uniform in diameter (~30 nm). Next, the prepared magnetic nanoparticles were adjusted to a concentration of 1 µM with DEPC treated water, mixed with 10 µM of K-ras G12V or G13D mutation-specific capture probe in the presence of 1-ethyl-3-(-3-dimethylaminopropyl) carbodiimide hydrochloride (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), and purified in a magnetic field. This was followed by redispersion in Tris-EDTA buffer and the addition of a cross-linker (50 µM of EDC) to react for 2 hours. Then, the magnetic nanoparticles were collected by magnetic separation and re-suspended in TE.
K-ras mutation detection
K-ras mutation detection was performed by fluorescent quantitative real-time PCR in a 20-µL reaction system containing 20 ng of sample DNA, 0.9 mmol/L of each primer, 0.9 mmol/L of inhibitor, and 0.20 mmol/L of nanoparticle probe on an ABI 7500 real-time PCR cycler. The PCR cycling parameters were pre-denaturation at 95 °C for 10 minutes and 40 cycles of 95 °C for 20 seconds and 60 °C for 45 seconds. The probe, primers, and blockers used were as follows: K-ras capture probe, 5'-CTCTATTGTTGGATCATATTCGTCCACAAAATGATTCTGAATTA-3'; G12V forward primer, 5'-ACTTGTGGTAGTTGGACCT-3' and reverse primer, 5'-TAACTTGAAACCCAAGGTAC-3'; blocker, CCTACGCCACCAGCT (with 4 pentabases); G13D forward primer, 5'-GTTCTAATATAGTCACATTTTCATTATTTTTATTATAAAGC-3' and reverse primer, 5'-GTCAAGGCACTCTTGCCTAGG-3'; blocker, CTTGCCTACGCCACCA (with 4 pentabases); β-actin forward primer, 5'-CTCCAT CCTGGCCTCGCTGT-3' and reverse primer, 5'-GCTGTCACCTTCACCGTTCC-3'. Samples with a Ct value of 21–37 were regarded as having a mutant allele, while samples with a Ct value of 15–20 were regarded as having a wild-type allele.
Statistical analysis
Statistical analyses were performed using SPSS 19.0 statistical software. The sensitivity, specificity, positive predictive value, and negative predictive value of the nanoprobe-based plasma and stool K-ras gene detection, alone or in combination, in the diagnosis of pancreatic cancer were calculated. Numerical data, which are expressed as mean ± standard deviation, were compared using the t-test. Categorical data, which are expressed as percentages, were compared using the χ2 test. Survival data were computed using the Kaplan-Meier method and compared using the Log-rank test. Paired dichotomous data were compared using the McNemar test. P values <0.05 were considered statistically significant.
Results
Nanoprobe-based K-ras G12V and G13D mutation detection in plasma and stool samples
Table 1 shows the K-ras G12V and G13D mutation status detected in stool and plasma samples from different groups of subjects. The detection rates of K-ras mutations in stool and plasma samples from patients with pancreatic cancer were 79.3% and 43.1%, respectively, and G12V was the major mutation type (40/46 and 21/25, respectively). The detection rates of K-ras mutations in stool and plasma samples from patients with pancreatic cancer were significantly higher than those from patients with benign pancreatic diseases (15.6% and 6.3%, respectively; P<0.05) and normal controls (0% for both; P<0.05). K-ras mutations were detected in both plasma and stool samples from 21 patients with pancreatic cancer and one patient with benign pancreatic disease.

Full table
Diagnostic value of nanoprobe-based plasma and stool K-ras mutation testing in patients with pancreatic cancer
As shown in Table 2, stool K-ras mutation status detected by nanoprobe-based assay had sensitivity and specificity of 79.3% and 84.4%, respectively, in the diagnosis of pancreatic cancer, and the corresponding values for plasma K-ras mutation status were 43.1% and 93.8%, respectively. When combining the K-ras mutation status in stool and plasma together, the sensitivity and specificity rose to 86.2% and 96.9%, respectively.
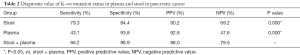
Full table
Relationship of plasma and stool K-ras mutation status detected by nanoprobe-based assay with the clinicopathological characteristics of pancreatic cancer
We next analyzed the relationship between nanoprobe-based plasma and stool K-ras mutation testing and the clinicopathological characteristics of pancreatic cancer, including gender, age, the presence of clinical symptoms, tumor location, tumor size, tumor differentiation, carbohydrate antigen 19-9 (CA19-9) level, carcinoembryonic antigen (CEA) level, and TNM stage. As shown in Table 3, the K-ras mutation status in stool had no significant association with these clinicopathological characteristics. However, K-ras mutation status in plasma was significantly associated with the TNM stage, although this had no significant association with other clinicopathological characteristics (Table 4).
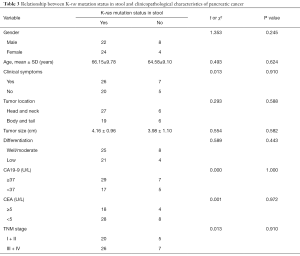
Full table
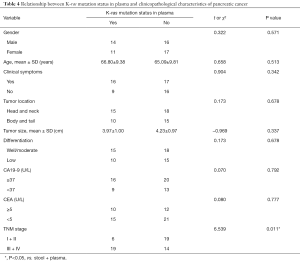
Full table
Relationship of plasma and stool K-ras mutation status detected by nanoprobe-based assay with the survival of patients with pancreatic cancer
All patients were followed by telephone or at the outpatient clinic. The duration of the follow-up period ranged from 2 to 26 months, with a mean value of 7.3 months. None of the patients were lost to follow-up. There was no significant difference in either the 6-month or 1-year survival rate between patients with and without K-ras mutations in stool (78.8% vs. 81.5%, P=0.884; 41.2% vs. 45.3%, P=0.698) (Figure 1A,B). However, the 1-year survival rate was significantly lower in patients with K-ras mutations in plasma than in those without (24.5% vs. 58.1%, P=0.030), although there was no significant difference in the 6-month survival rate (73.6% vs. 83.7%, P=0.388) (Figure 1C,D).
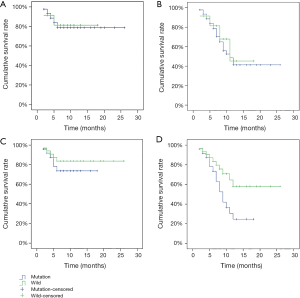
Discussion
In the present study, we assessed the diagnostic value of the plasma and stool K-ras mutation status detected by a nanoprobe-based assay, and analyzed the association of K-ras mutations in plasma and stool samples with clinicopathological characteristics and survival in patients with pancreatic cancer. It was found that K-ras mutations could be detected in a significant number of patients. However, these mutations could only be detected in a minority of patients with benign pancreatic diseases. The combination of K-ras mutation status in stool and plasma resulted in satisfactory sensitivity and specificity for the diagnosis of pancreatic cancer (86.2% and 96.9%, respectively). In addition, it was noted that the K-ras mutation status in plasma was significantly associated with the TNM stage and 1-year survival, although this is not the case for K-ras mutation status in stool.
Since plasma DNA, in which K-ras mutations were detected, is present at extremely low levels (19), their detection requires a highly sensitive system. The nanoprobe-based assay system used in the present study combines the use of fluorescent nanoprobes and the technique of real-time PCR. On one hand, nanoparticles have a high surface area-to-volume ratio, which allows for more target molecules to be attached to a nanoparticle, greatly improving the diagnostic sensitivity (17,18). On the other hand, real-time PCR can produce quantitative data that spans 7–8 log orders of magnitude, and thereby has an extremely high sensitivity (20). Therefore, the investigators consider that the combination of both can meet the requirement of high sensitivity in detecting a target mutation from trace amounts of DNA.
The reported sensitivities of stool K-ras mutation detection for the diagnosis of pancreatic cancer vary greatly from 0% to 80%, and the specificities are lower than 70% (21). In the present study, stool K-ras mutation testing had a sensitivity of 79.3% and a specificity of 84.4% in the diagnosis of pancreatic cancer, indicating a moderate diagnostic accuracy. Thus, nanoprobe-based stool K-ras mutation testing alone can be used for the screening of pancreatic cancer. In contrast, plasma K-ras mutation detection had a very low sensitivity of 43.1% and a high specificity of 93.8%. This result is similar to the finding of a previous meta-analysis, which reported that plasma K-ras mutation testing had a pooled sensitivity and specificity of 37% and 91%, respectively (22), suggesting that plasma K-ras mutation testing alone, due to its low sensitivity, is only suitable for auxiliary diagnosis to improve diagnostic accuracy. However, when plasma and stool K-ras mutations were simultaneously detected, the sensitivity and specificity rose to 86.2% and 96.9%, respectively.
K-ras mutations could be detected in stool and plasma samples from a small portion of patients with benign pancreatic diseases (15.6% and 6.3%, respectively). This result suggests that the sensitivity and specificity of nanoprobe-based K-ras mutation testing will inevitably decrease in clinical settings, where the differential diagnosis between pancreatic cancer and benign pancreatic diseases is common. However, considering that benign pancreatic diseases (especially pancreatic epithelial cell hyperplasia) that carry K-ras alterations may be a premalignant condition (11), the detection of K-ras mutations in stool and plasma may identify patients at risk of developing pancreatic cancer.
Many studies have assessed the significance of K-ras mutation status in the prediction of prognosis in patients with pancreatic cancer. However, these results are in conflict with the use of K-ras mutation status as a prognostic factor for pancreatic cancer (10). Nonetheless, there appears to be a consensus that K-ras mutation status in plasma predicts a worse survival outcome in patients with pancreatic cancer (10,23,24), although survival might not differ by K-ras mutations in tissue DNA (25,26). In agreement with these findings, the present study revealed that plasma, not stool, K-ras mutation status was significantly associated with the 1-year survival rate of patients with pancreatic cancer (24.5% vs. 58.1%). Although the 6-month survival rate did not significantly differ by plasma K-ras mutation status, this exhibited a trend towards this (73.6% vs. 83.7%), which may be due to the small sample size of the study. The discrepancy in the prognostic significance of plasma and stool K-ras mutation status may be explained by the origin of plasma DNA. It has been hypothesized that plasma DNA might originate from tumor cells detached from metastases (24), and the detection of K-ras mutations in plasma may suggest that metastases have occurred. Thus, it is not surprising that plasma K-ras mutation status is associated with poorer prognosis.
Since the clinicopathological characteristics of pancreatic cancer often correlate with its prognosis, the relationship of plasma and stool K-ras mutation status with the clinicopathological characteristics of pancreatic cancer was also analyzed. Consistent with a previous finding that K-ras mutation status did not correlate with any of the clinicopathological parameters (24), no significant associations were detected, except that K-ras mutation status in plasma was significantly associated with the TNM stage. Furthermore, a previous study has noted that the presence of K-ras mutations in peripheral blood might reflect different tumor stages (14). This result can also be explained by the hypothesis that the detection of K-ras mutations in plasma suggests the presence of metastases (24), which may upgrade the tumor stage.
The present study has several major limitations. First, as mentioned above, the sample size was small, which may result in some insignificant associations. Second, the retrospective nature of the study may have caused errors and bias inherent to such kind of study. Third, the follow-up period was relatively short. Finally, false positives and false negatives associated with nanoprobe-based assay may lead to the possibility of failing to fully detect the true relationship of K-ras mutation status with the prognosis and clinicopathological characteristics of pancreatic cancer. Future studies are warranted to carefully address these issues.
In conclusion, we have developed a novel fluorescent nanoprobe-based assay for detecting K-ras mutations in plasma and stool samples from patients with pancreatic cancer. This assay exhibits satisfactory sensitivity and specificity when plasma and stool samples are simultaneously detected. The nanoprobe-based detection of plasma K-ras mutation status is associated with survival and tumor stage in patients with pancreatic cancer, and can therefore serve as an auxiliary tool for prognostic evaluation.
Acknowledgments
Funding: This study was supported by the Public Welfare Technology Research and Social Development Project of the Department of Science and Technology of Zhejiang Province (No. 2014C33139) and the Social Development Research and Demonstration Application Project of the Bureau of Science and Technology of Jiaxing City (No. 2016AY23057).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.07.07). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study protocol was approved by the Ethics Committee of the Second Affiliated Hospital of Jiaxing University (No. L2015-28). Informed consent was obtained from all participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7-30. [Crossref] [PubMed]
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Vincent A, Herman J, Schulick R, et al. Pancreatic cancer. Lancet 2011;378:607-20. [Crossref] [PubMed]
- Dunne RF, Hezel AF. Genetics and biology of pancreatic ductal adenocarcinoma. Hematol Oncol Clin North Am 2015;29:595-608. [Crossref] [PubMed]
- Buscail L, Bournet B, Dufresne M, et al. Advance in the biology of pancreatic of cancer. Bull Cancer 2015;102:S53-61. [Crossref] [PubMed]
- Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer 2011;11:761-74. [Crossref] [PubMed]
- Almoguera C, Shibata D, Forrester K, et al. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell 1988;53:549-54. [Crossref] [PubMed]
- Logsdon CD, Lu W. The significance of Ras activity in pancreatic cancer initiation. Int J Biol Sci 2016;12:338-46. [Crossref] [PubMed]
- Yang J, Li J, Zhu R, et al. K-ras mutational status in cytohistological tissue as a molecular marker for the diagnosis of pancreatic cancer: a systematic review and meta-analysis. Dis Markers 2014;2014:573783 [Crossref] [PubMed]
- Tao LY, Zhang LF, Xiu DR, et al. Prognostic significance of K-ras mutations in pancreatic cancer: a meta-analysis. World J Surg Oncol 2016;14:146. [Crossref] [PubMed]
- Mulcahy H, Farthing MJ. Diagnosis of pancreatico-biliary malignancy: detection of gene mutations in plasma and stool. Ann Oncol 1999;10:114-7. [Crossref] [PubMed]
- Dobre M, Comănescu M, Arsene D, et al. K-ras gene mutation status in colorectal cancer: comparative analysis of pyrosequencing and PCR-RFLP. Rom J Morphol Embryol 2013;54:567-74. [PubMed]
- Oltedal S, Gilje B, Kørner H, et al. Detection of occult metastases in sentinel lymph nodes from colon cancer patients by K-ras mutation peptide nucleic acid clamp PCR. Ann Surg 2010;251:1087-91. [Crossref] [PubMed]
- Däbritz J, Preston R, Hänfler J, et al. Follow-up study of K-ras mutations in the plasma of patients with pancreatic cancer: correlation with clinical features and carbohydrate antigen 19-9. Pancreas 2009;38:534-41. [Crossref] [PubMed]
- Rogosnitzky M, Danks R. Validation of blood testing for K-ras mutations in colorectal and pancreatic cancer. Anticancer Res 2010;30:2943-7. [PubMed]
- Jianrong C, Yuqing M, Nongyue H, et al. Nanotechnology and biosensors. Biotechnol Adv 2004;22:505-18. [Crossref] [PubMed]
- Chi X, Huang D, Zhao Z, et al. Nanoprobes for in vitro diagnostics of cancer and infectious diseases. Biomaterials 2012;33:189-206. [Crossref] [PubMed]
- Jain KK. Nanodiagnostics: application of nanotechnology in molecular diagnostics. Expert Rev Mol Diagn 2003;3:153-61. [Crossref] [PubMed]
- Esmaeilsabzali H, Beischlag TV, Cox ME, et al. Detection and isolation of circulating tumor cells: principles and methods. Biotechnol Adv 2013;31:1063-84. [Crossref] [PubMed]
- Wong ML, Medrano JF. Real-time PCR for mRNA quantitation. Biotechniques 2005;39:75-85. [Crossref] [PubMed]
- Haug U, Wente MN, Seiler CM, et al. Stool testing for the early detection of pancreatic cancer: rationale and current evidence. Expert Rev Mol Diagn 2008;8:753-9. Erratum in: Expert Rev Mol Diagn 2012;12:110 Jesnowski, Ralf [corrected to Jesenofsky, Ralf]. [Crossref] [PubMed]
- Liu SL, Chen G, Zhao YP, et al. Diagnostic accuracy of K-ras mutation for pancreatic carcinoma: a meta-analysis. Hepatobiliary Pancreat Dis Int 2013;12:458-64. [Crossref] [PubMed]
- Chen H, Tu H, Meng ZQ, et al. K-ras mutational status predicts poor prognosis in unresectable pancreatic cancer. Eur J Surg Oncol 2010;36:657-62. [Crossref] [PubMed]
- Singh N, Gupta S, Pandey RM, et al. High levels of cell-free circulating nucleic acids in pancreatic cancer are associated with vascular encasement, metastasis and poor survival. Cancer Invest 2015;33:78-85. [Crossref] [PubMed]
- Li T, Zheng Y, Sun H, et al. K-Ras mutation detection in liquid biopsy and tumor tissue as prognostic biomarker in patients with pancreatic cancer: a systematic review with meta-analysis. Med Oncol 2016;33:61. [Crossref] [PubMed]
- Kinugasa H, Nouso K, Miyahara K, et al. Detection of K-ras gene mutation by liquid biopsy in patients with pancreatic cancer. Cancer 2015;121:2271-80. [Crossref] [PubMed]


