
Identification of potential therapeutic targets in hepatocellular carcinoma using an integrated bioinformatics approach
Introduction
Hepatocellular carcinoma (HCC), a heterogeneous cancer, is one of the most common malignant tumors and the main cause for cancer-related death worldwide (1). Major risk factors for HCC include aflatoxin exposure, tobacco use, nonalcoholic fatty liver disease, metabolic syndrome, and various carcinogens. Hepatitis B and C viral infection also cause HCC, although this is more common in sub-Saharan Africa and Eastern Asia (2,3). Surgical resection is currently the first-line treatment for HCC, specifically liver resection, liver transplantation, and local ablation (4). Other therapeutic options include systemic treatments that are individualized for each patient (5). Unfortunately, not all therapies are effective, and frequent tumor recurrence and poor prognosis are common problems associated with HCC, particularly when it is diagnosed at an advanced stage. Thus, there is a need to better understand the molecular pathogenesis of HCC and explore novel therapies.
Recent research has been undertaken to better understand molecules and pathways related to tumorigenesis, development and progression of HCC. Li et al. (6) reported that the single-nucleotide polymorphism (SNP) rs2380585 in the ADAMTS5 gene contributes to susceptibility to HCC in a Chinese Han population. Furthermore, overexpression of the general transcription factor IIB (GTF2B) may contribute to HCC pathogenesis via cell proliferation (7). Zeng et al. (8) revealed that the activation of transcription factor SALL4 enhances spheroid formation and invasiveness of a HCC subtype with stem cell features. Murakami and colleagues (9) found that some miRNAs are overexpressed in HCC compared to adjacent non-tumorous tissues and are linked to differentiation of HCC. Cao et al. (10) reported that the overexpression of DDB1 and CUL4-associated factor 13, which regulate cell cycle progression, is associated with poor survival in HCC. Previous studies (11,12) have also identified molecular targets, including epidermal growth factor receptor, platelet-derived growth factor receptor, histone deacetylases, fibroblast growth factor, and vascular endothelial growth factor receptor, which play critical roles in the development, progression, and metastasis of HCC. Furthermore, research investigations on pathway dysregulation may improve our understanding of HCC pathobiology. A meta-analysis (13) of gene expression profiles has identified three robust HCC molecular subclasses, which includes the activation of the WNT signaling pathway. The aberrant activation of the Hedgehog, RAS/MAPK, PI3K/Akt, and tyrosine kinase receptor-related signaling pathways have also been associated with HCC (14). However, information on the underlying molecular mechanisms of HCC pathogenesis is limited.
We report data from three microarray datasets downloaded from GEO database and the identification of differentially expressed genes (DEGs) between HCC and adjacent non-tumorous tissues. GO functional and KEGG pathway enrichment analysis were conducted to analyze the potential functions of the identified DEGs. In addition, protein-protein interaction (PPI) network and sub-network analyses were performed to screen for hub genes. These analyses were conducted to identify potential biomarkers for HCC and to elucidate its underlying pathological mechanisms.
Methods
Microarray data
In the present study, three gene expression datasets, namely, GSE36376 (15), GSE45267 (Wang et al., personal communication), GSE51401 (Sun et al., personal communication) were downloaded from the National Center of Biotechnology Information (NCBI) Gene Expression Omnibus (GEO available online: https://www.ncbi.nlm.nih.gov/geo/) database, which is an international public functional genomic data repository. GSE36376 is based on the GPL10558 platform (Illumina HumanHT-12 V4.0 expression beadchip). GSE45267 and GSE51401 are based on the same platform of GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array. The present study included 584 samples (318 HCC and 266 adjacent non-tumorous tissues). Specifically, GSE36376 included 240 tumor and 193 non-cancerous liver tissues specimens containing no necrosis or hemorrhage from HCC patients who were treated with surgical resection or liver transplantation. None of the patients received preoperative chemotherapy. GSE45267 comprised 48 tumor and 39 non-cancerous specimens from 61 patients, HCC tissues were collected from 16 young HCC and 32 elder HCC patients. GSE51401 consisted of 30 tumor and 34 non-cancerous specimens, which were collected immediately after removal from patients with pathologically proven HCC and positive alpha-fetoprotein (AFP), then washed 3 times by phosphate buffer solution (PBS). All samples were obtained from patients with HCC. This study used gene expression data downloaded from the public database, thus no patient consent or ethics committee approval were necessary.
Data preprocessing
Series matrix files and annotation information of the corresponding platform for the three gene expression microarray datasets were downloaded. Value distributions of the three datasets were also downloaded from the GEO database, which were shown in (Figures S1-S3); median-centered values were indicative that the data were normalized and cross-comparable. R packages were used for data preprocessing, which consisted of data normalization and background correction. In combination with annotation information files for the platform of GPL10558 and GPL570, probe IDs were transformed into gene names. Gene expression values were calculated and used in the data analysis.
DEG screening
The DEGs in the three microarray datasets (GSE36376, GSE45267, and GSE51401) from HCC specimens were compared to those of non-cancerous specimens using the limma package in R language. The threshold for the selection of significant DEGs was |log2 fold change (log2FC)| >1 and an adjusted P<0.01. Additionally, the intersection of DEGs among the three microarray datasets was screened using Venny2.1.0 (http://bioinfogp.cnb.csic.es/tools/venny/index.html). The overlapping DEGs were represented using a Volcano plot using ggplot2 in R.
GO annotation and KEGG pathway analysis of DEGs
Functional annotation of the identified DEGs using GO analysis indicated the enrichment of three categories, namely, cellular component (CC), biological process (BP), and molecular function (MF). KEGG pathway analysis provided information on biological chemicals, genomes, and enzymatic pathways (16). To analyze the function of common DEGs, GO annotation and KEGG pathway enrichment analysis were performed using R (version: 3.4.1) language. The threshold for significantly enriched DEGs was P<0.01.
Construction of the PPI network and sub-network
The Search Tool for the Retrieval of Interacting Genes (STRING; https://string-db.org/) online database (17), which encompasses various organisms, was used to predict PPIs, including direct (physical) and indirect (functional) interactions. Interactions among the identified DEGs were analyzed by mapping with STRING (version: 10.5), and a combined score ≥0.4 was used as criterion. Cytoscape (version:3.5.1; http://cytoscape.org/), a visualization software, was used to construct the PPI network and sub-network based on the data generated using STRING. The proteins and their interactions were shown using nodes and edges, respectively. Nodes with more connectivity and combined scores were considered hub genes.
Results
DEG analysis of integrated microarray datasets
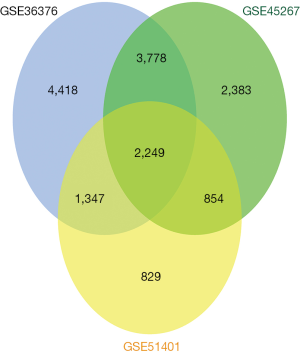
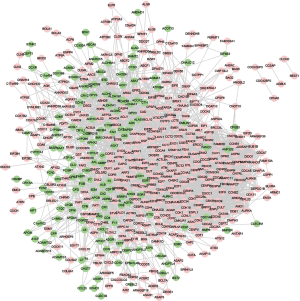
Based on the aforementioned selection criteria, the three microarray datasets were downloaded from GEO, and the expression datasets for GSE36376, GSE45267, and GSE51401 were used to extract DEGs (Figure 1). Of these DEGs, 2,249 overlapping DEGs were used for subsequent analysis of HCC and non-cancerous tissues and their corresponding distribution (Figures 1,2).

Functional and pathway enrichment analyses
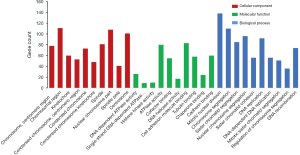
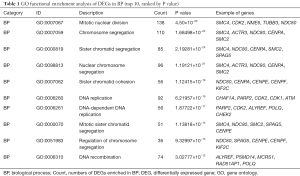
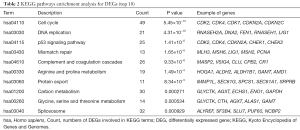
GO and KEGG analyses were performed to further investigate common DEGs that are associated with HCC. GO analyses of DEGs, identified three functional categories (BP, MF and CC) (Figure 3). Functional enrichment analysis involving the BP category indicated that the identified DEGs are predominantly associated with mitotic nuclear division, chromosome segregation, and sister chromatid segregation (Table 1). KEGG pathway analysis showed that the functional categories cell cycle, DNA replication, and p53 signaling pathway were significantly enriched with the common DEGs (Table 2).
Full table
Full table
PPI network and sub-network analysis
The PPI network of the common DEGs consisted of 565 nodes and 4,223 edges (Figure 4). Among these genes, cyclin B1 (CCNB1) showed the greatest degree in the PPI network, suggesting that CCNB1 plays a key role in HCC development. To learn more about the common DEGs, genes with combined scores of ≥0.999 were selected in the construction of a PPI sub-network (Figure 5). The hub nodes in the PPI sub-network included cyclin dependent kinase 2 (CDK2), cyclin B1 (CCNB1), and cell division cycle 20 (CDC20).
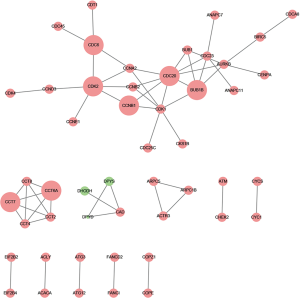
Discussion
HCC is a complex malignancy involving various genetic and epigenetic alterations. However, its underlying pathology is not well understood. The present study conducted an integrated bioinformatics analysis of three datasets from GEO and identified 2,249 DEGs that were observed in both HCC and non-cancerous tissues. Among these DEGs, 1,735 were upregulated and 514 were downregulated. GO and KEGG enrichment analyses showed that the functional categories of cell cycle, DNA replication, and the p53 signaling pathway were the most highly enriched. PPI network and sub-network analysis identified CDK2, CCNB1 and CDC20 as hub nodes. Taken together, these findings indicate that these specific genes and pathways are involved in HCC progression.
According to the STRING online database, CDK2, CCNB1 and CDC20 are all involved in cell cycle and oocyte meiosis, and they have interactions with each other, indicating that these genes might also implicate in HCC through interacting with each other. It has been reported that the DEGs between liver cancer tissues and non-cancerous tissues were enriched in cell cycle and oocyte meiosis (18), which were the significantly dysregulated pathways in HCC (19). The eukaryotic cell cycle contains the alternation between chromosome segregation and DNA replication, which is driven by the Cyclin-Cdk family of protein kinases (20). Individual cyclin functions at different stages of cell cycle via binding and activating the corresponding CDK protein (21). Oocyte meiosis occurs in the absence of any appreciable transcription. During meiosis, a single round of DNA replication is followed by two rounds of chromosome segregation, which are called meiosis I and meiosis II. CDC20 is referred to the microtubule function in the process of meiosis; specifically, CDC20 is essential for the anaphase onset of the meiosis I but not the meiosis II in mouse oocytes (22). A study demonstrated that the activity of CDK2 was required for oocyte meiosis (23). CCNB1 is required for cells to enter mitosis and its spatial localization plays an important role in triggering mitosis at the correct time (24). Thus, these genes have interaction with each other by the pathways.
CDK2 is a CDK that is involved in DNA replication, histone synthesis, centrosome duplication, and the cell cycle. Among these processes, CDK2 is a vital regulator of eukaryotic cell cycle progression, which strictly controls cell proliferation, growth, and survival. Cell cycle dysregulation may cause uncontrolled proliferation, thereby resulting in malignancies (25,26). CDK2 regulates the G1/S phase by binding to positive regulators such as cyclin E and cyclin A (27,28). The CDK2/cyclin A complex play a major role in regulating the progression of the S-phase, whereas the CDK2/cyclin E complex is required for the G1 to S-phase transition. However, aberrant expression of CDK2 may lead to uncontrolled cell proliferation (29), which is associated with tumor formation. CDK2 has been correlated to cancer (30). Kohzato et al. (31) reported that CDK2 and cyclin E are both overexpressed in HCC; however, the overexpression of CDK2 has been strongly associated with poorly differentiated HCC. In our study, CDK2 was identified as a hub gene in the PPI network and sub-network and was significantly enriched the cell cycle functional category, suggesting that CDK2 contributes to the pathogenesis of HCC.
CCNB1, a member of the highly conserved cyclin B family, is expressed in nearly all human body tissues (21). CCNB1 controls the G2/M phase transition in the cell cycle, and its deregulation can result in uncontrolled cell cycle progression and tumorigenesis. The CCNB1 has also been correlated to various malignancies (32). Song et al. (33) reported that CCNB1 induces cancer cell invasiveness and metastasis in esophageal squamous cell carcinoma. Matthess and colleagues (34) indicated that CCNB1 controls Fas-mediated apoptosis by regulating caspase-8 activity, which contributes to tumorigenesis. Gomez et al. (35) showed that CCNB1 is implicated in chemoresistance in prostate cancer cells. Zhao et al. (36) revealed that the upregulation of CCNB1 is strongly associated with pituitary adenomas invasiveness, whereas its downregulation decreases cell proliferation (37). Other studies have suggested that CCNB1 is associated with poor prognosis in patients with tumors (38). Previous studies (39) have also revealed that CCNB1 is correlated with HCC that may potentially be utilized as a candidate biomarker or a therapeutic target. The present study conducted functional and pathway enrichment analyses of DEGs in HCC, which further support previous findings that CCNB1 is correlated with the cell cycle and the p53 signaling pathway. Thus, CCNB1 may play a pivotal role in tumorigenesis and progression of HCC.
Abnormal cell-cycle checkpoints may result in cancer. CDC20, which is a regulatory protein in cell cycle checkpoints (40), is required for nuclear movement before anaphase and chromosome separation by interacting with other proteins. It activates the anaphase-promoting complex at metaphase and is also involved in mitotic exit (41). In our study, GO analysis of DEGs in BP showed that the functional categories of mitotic nuclear division, chromosome segregation, sister chromatid segregation, nuclear chromosome segregation, sister chromatid cohesion are enriched with CDC20. Pathway analysis revealed that the cell cycle pathway and spliceosome are enriched with CDC20. Furthermore, PPI network and sub-network analyses demonstrated that CDC20 is a hub gene. CDC20 plays a significant role in the initiation and progression of various tumors (42). Previous bioinformatics analysis (18) has indicated that CDC20 may be associated with carcinogenesis, invasion, or recurrence of HCC. Li et al. (42) revealed that CDC20 is upregulated in 68% of HCC patients and is positively correlated with clinicopathological parameters such as gender, TNM stage, and tumor differentiation. Furthermore, silencing of CDC20 expression using siRNA delays HCC progression, decreases cell proliferation, and induces entry into the G2/M cell cycle phase (42). Huang et al. (43) reported that inhibiting tumor cell growth (including apoptosis-resistant, SAC-defective, and slippage-prone cancer cells) by knocking down CDC20 blocks downstream mitotic exit. Thus, CDC20 may be potentially used as a biomarker and a novel therapeutic target for HCC.
What’s more, the result of data analysis should be connected with clinic research. The report showed that CCNB1 was highly expressed in HCC (44). The study by Weng et al. (45) showed that CCNB1 positivity was observed in 80% of the recurrent HCC samples related to the hepatitis B virus infection. Furthermore, high or low expression of CCNB1 had no significant correlations with any single clinicopathological characteristics, including sex, age, AFP, tumor number, histopathological grading, or liver cirrhosis. However, elevated expressed CCNB1 was connected with earlier recurrence in HCC patients after surgical resection, and its overexpression could be a prognostic factor for recurrence-free survival. CDC20 is one of the regulators of cell cycle checkpoints. The research (42) demonstrated that the expression of CDC20 was abnormal higher in primary HCC samples than the non-cancerous samples from the same patients. Upon spearman correlation analysis, overexpression of CDC20 was closely correlated with gender, advanced TNM staging, poorer tumor differentiation, and higher expression level of p53. Nevertheless, increased CDC20 expression revealed no statistically significant correlation with other clinicopathological features, including age, HBV infection, hepatic cirrhosis, serum AFP level, tumor size, vascular invasion and intra/extra hepatic metastasis. CDK2 is the key protein for the transition of cell cycle G1/S phase. CDK2 is activated in over 80% of HCC cases. Whereas, no significant association was found between the degree of CDK activation and the clinical parameters of HCC samples, including cellular differentiation, tumour size, tumour encapsulation, venous permeation, and microsatellite (46). The aforementioned results by our study showed that CCNB1, CDC20 and CDK2 were the hub genes from the network. However, they were needed to be correlated with the clinical validation, which would be carried out in our further researches.
Conclusions
We identified DEGs in HCC tissues, and CDK2, CCNB1, and CDC20 were classified as hub genes that play a predominate role in HCC tumorigenesis. These DEGs are associated with major biological pathways such as cell cycle, DNA replication, and the p53 signaling pathway. These genes may be potentially utilized as therapeutic targets and prognostic biomarkers for HCC. Also, our data improves our understanding of the underlying mechanisms of HCC, although further investigations are warranted.


Acknowledgments
We would like to thank LetPub for providing linguistic assistance during the preparation of this manuscript.
Funding: The National Natural Science Foundation of China (Grant number 81760851) and the Foundation of Guangxi University of Chinese Medicine (Grant number P2012116) supported this study.
Footnote
Conflicts of Interest: ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.06.04). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study used gene expression data downloaded from the public database, thus no patient consent or ethics committee approval were necessary.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- El-Serag HB, Kanwal F. Epidemiology of Hepatocellular Carcinoma in the United States: Where Are We? Where Do We Go? Hepatology 2014;60:1767. [Crossref] [PubMed]
- Dhanasekaran R, Limaye A, Cabrera R. Hepatocellular carcinoma: current trends in worldwide epidemiology, risk factors, diagnosis, and therapeutics. Hepat Med 2012;4:19-37. [PubMed]
- El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 2012;142:1264. [Crossref] [PubMed]
- Llovet JM, Bruix J. Novel advancements in the management of hepatocellular carcinoma in 2008 ☆. J Hepatol 2008;48:S20. [Crossref] [PubMed]
- Chan SL, Yeo W. Development of systemic therapy for hepatocellular carcinoma at 2013: Updates and insights. World J Gastroenterol 2014;20:3135-45. [Crossref] [PubMed]
- Li C, Xiong Y, Zhong Z, et al. Association Between a Variant in ADAMTS5 and the Susceptibility to Hepatocellular Carcinoma in a Chinese Han Population. Cell Biochem Biophys 2015;72:221-5. [Crossref] [PubMed]
- Li L, Zhang A, Cao X, et al. General transcription factor IIb overexpression and a potential link to proliferation in human hepatocellular carcinoma. Pathol Oncol Res 2013;19:195-203. [Crossref] [PubMed]
- Zeng SS, Yamashita T, Kondo M, et al. The transcription factor SALL4 regulates stemness of EpCAM-positive hepatocellular carcinoma. J Hepatol 2014;60:127-34. [Crossref] [PubMed]
- Murakami Y, Yasuda T, Saigo K, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene 2006;25:2537-45. [Crossref] [PubMed]
- Cao J, Hou P, Chen J, et al. The overexpression and prognostic role of DCAF13 in hepatocellular carcinoma. Tumour Biol 2017;39:1010428317705753 [Crossref] [PubMed]
- Chuma M, Terashita K, Sakamoto N. New molecularly targeted therapies against advanced hepatocellular carcinoma: From molecular pathogenesis to clinical trials and future directions. Hepatol Res 2015;45:E1-11. [Crossref] [PubMed]
- Lee YH, Seo D, Choi KJ, et al. Antitumor effects in hepatocarcinoma of isoform-selective inhibition of HDAC2. Cancer Res 2014;74:4752-61. [Crossref] [PubMed]
- Hoshida Y, Nijman SM, Kobayashi M, et al. Integrative Transcriptome Analysis Reveals Common Molecular Subclasses of Human Hepatocellular Carcinoma. Cancer Res 2009;69:7385-92. [Crossref] [PubMed]
- Villanueva A, Newell P, Chiang DY, et al. Genomics and Signaling Pathways in Hepatocellular Carcinoma. Semin Liver Dis 2007;27:55-76. [Crossref] [PubMed]
- Lim HY, Sohn I, Deng S, et al. Prediction of Disease-free Survival in Hepatocellular Carcinoma by Gene Expression Profiling. Ann Surg Oncol 2013;20:3747-53. [Crossref] [PubMed]
- Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucl Acids Res 2000;28:27-30. [Crossref] [PubMed]
- Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 2017;45:D362-8. [Crossref] [PubMed]
- Li L, Lei Q, Zhang S, et al. Screening and identification of key biomarkers in hepatocellular carcinoma: Evidence from bioinformatic analysis. Oncol Rep 2017;38:2607-18. [Crossref] [PubMed]
- Jin B, Wang W, Du G, et al. Identifying hub genes and dysregulated pathways in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci 2015;19:592. [PubMed]
- Morgan DO. The Cell Cycle: Principles of Control. Primers in Biology. London: New Science, 2006.
- Miyazaki T, Arai S. Two distinct controls of mitotic cdk1/cyclin B1 activity requisite for cell growth prior to cell division. Cell Cycle 2007;6:1419-25. [Crossref] [PubMed]
- Yin S, Liu JH, Ai JS, et al. Cdc20 is required for the anaphase onset of the first meiosis but not the second meiosis in mouse oocytes. Cell Cycle 2007;6:2990-2. [Crossref] [PubMed]
- Gabrielli BG, Roy LM, Maller JL. Requirement for Cdk2 in cytostatic factor-mediated metaphase II arrest. Science 1993;259:1766-9. [Crossref] [PubMed]
- Strauss B, Harrison A, Coelho PA, et al. Cyclin B1 is essential for mitosis in mouse embryos, and its nuclear export sets the time for mitosis. J Cell Biol 2018;217:179-93. [Crossref] [PubMed]
- Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer 2009;9:153-66. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646. [Crossref] [PubMed]
- Sherr CJ. G1 phase progression: Cycling on cue. Cell 1994;79:551-5. [Crossref] [PubMed]
- Lim S, Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development 2013;140:3079-93. [Crossref] [PubMed]
- Sakurikar N, Thompson R, Montano R, et al. A subset of cancer cell lines is acutely sensitive to the Chk1 inhibitor MK-8776 as monotherapy due to CDK2 activation in S phase. Oncotarget 2016;7:1380-94. [Crossref] [PubMed]
- Kaklamani VG, Jeruss JS, Hughes E, et al. Phase II neoadjuvant clinical trial of carboplatin and eribulin in women with triple negative early-stage breast cancer (NCT01372579). Breast Cancer Res Treat 2015;151:629. [Crossref] [PubMed]
- Kohzato N, Dong Y, Sui L, et al. Overexpression of cyclin E and cyclin-dependent kinase 2 is correlated with development of hepatocellular carcinomas. Hepatol Res 2001;21:27-39. [Crossref] [PubMed]
- Fang Y, Yu H, Liang X, et al. Chk1-induced CCNB1 overexpression promotes cell proliferation and tumor growth in human colorectal cancer. Cancer Biol Ther 2014;15:1268-79. [Crossref] [PubMed]
- Song Y, Zhao C, Dong L, et al. Overexpression of cyclin B1 in human esophageal squamous cell carcinoma cells induces tumor cell invasive growth and metastasis. Carcinogenesis 2008;29:307-15. [Crossref] [PubMed]
- Matthess Y, Raab M, Sanhaji M, et al. Cdk1/cyclin B1 controls Fas-mediated apoptosis by regulating caspase-8 activity. Mol Cell Biol 2010;30:5726-40. [Crossref] [PubMed]
- Gomez LA, de Las Pozas A, Reiner T, et al. Increased expression of cyclin B1 sensitizes prostate cancer cells to apoptosis induced by chemotherapy. Mol Cancer Ther 2007;6:1534-43. [Crossref] [PubMed]
- Zhao P, Zhang P, Hu W, et al. Upregulation of cyclin B1 plays potential roles in the invasiveness of pituitary adenomas. J Clin Neurosci 2017;43:267-73. [Crossref] [PubMed]
- Müssnich P, Raverot G, Jaffrainrea ML, et al. Downregulation of miR-410 targeting the cyclin B1 gene plays a role in pituitary gonadotroph tumors. Cell Cycle 2015;14:2590-7. [Crossref] [PubMed]
- Huang T, Chen S, Han H, et al. Expression of Hsp90α and cyclin B1 were related to prognosis of esophageal squamous cell carcinoma and keratin pearl formation. Int J Clin Exp Pathol 2014;7:1544-52. [PubMed]
- Calvisi DF, Simile MM, Ladu S, et al. Activation of v-Myb avian myeloblastosis viral oncogene homolog-like2 (MYBL2)-LIN9 complex contributes to human hepatocarcinogenesis and identifies a subset of hepatocellular carcinoma with mutant p53. Hepatology 2011;53:1226-36. [Crossref] [PubMed]
- Weinstein J, Jacobsen FW, Hsu-Chen J, et al. A novel mammalian protein, p55CDC, present in dividing cells is associated with protein kinase activity and has homology to the Saccharomyces cerevisiae cell division cycle proteins Cdc20 and Cdc4. Mol Cell Biol 1994;14:3350-63. [Crossref] [PubMed]
- Pines J. Mitosis: a matter of getting rid of the right protein at the right time. Trends Cell Biol 2006;16:55-63. [Crossref] [PubMed]
- Li J, Gao JZ, Du JL, et al. Increased CDC20 expression is associated with development and progression of hepatocellular carcinoma. Int J Oncol 2014;45:1547-55. [Crossref] [PubMed]
- Huang HC, Shi J, Orth JD, et al. Evidence that Mitotic Exit Is a Better Cancer Therapeutic Target Than Spindle Assembly. Cancer Cell 2009;16:347-58.
- Covini G, Chan EK, Nishioka M, et al. Immune response to cyclin B1 in hepatocellular carcinoma. Hepatology 1997;25:75-80. [Crossref] [PubMed]
- Weng L, Du J, Zhou Q, et al. Identification of cyclin B1 and Sec62 as biomarkers for recurrence in patients with HBV-related hepatocellular carcinoma after surgical resection. Mol Cancer 2012;11:39. [Crossref] [PubMed]
- Li KK, Ng IO, Fan ST, et al. Activation of cyclin-dependent kinases CDC2 and CDK2 in hepatocellular carcinoma. Liver 2002;22:259-68. [Crossref] [PubMed]


